Enabling label-free detection of surface components in multicomponent samples with ultrahigh spatial resolution, tip-enhanced Raman spectroscopy makes it suitable for studying self-assembled monolayers, protein and nucleic acid chain composition, and cell surface interactions.
MARUDA SHANMUGASUNDARAM AND FRAN ADAR, HORIBA INSTRUMENTS INC.
Despite being able to provide fingerprint information in biology, medicine and other fields, Raman spectroscopy suffers from a signal-to-noise ratio that is limited by the inherent weakness of Raman scattering and a spatial resolution that is diffraction-limited (~0.5λ). This weak Raman signal-to-noise ratio can be overcome by surface-enhanced Raman spectroscopy (SERS), which relies on surface plasmon oscillations of metal particles in contact with the material1. However, it is often desirable to advance spatial resolution beyond the diffraction limit into the nanoscale dimension, which can provide fingerprint information with potentially single-molecule resolution. This spatial resolution enhancement — achievable through a technique called tip-enhanced Raman spectroscopy (TERS) — invites a broad range of applications in biology, such as the study of self-assembled monolayers, protein and nucleic acid chain composition, and cell surface interactions.
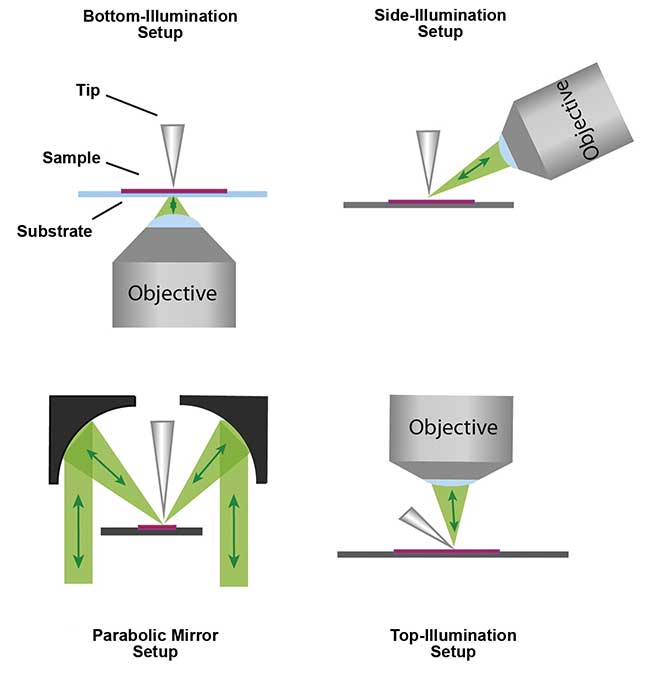
Figure 1. Illustration of different configurations for laser illumination of the tip-sample junction and
signal collection for tip-enhanced Raman spectroscopy. Adapted with permission from G. Sharma, et al. (July 2015). Tip-enhanced Raman scattering — Targeting structure-specific characterization for biomedical samples. Adv Drug Deliv Rev, Vol. 89, pp. 42-56. Copyright 2015 Elsevier.
TERS uses a metallized scanning probe microscope (SPM) tip to provide chemical information with the specificity and sensitivity of SERS but with nanoscale spatial resolution. Ever since the innovation of SPM techniques such as atomic force microscopy (AFM) and scanning tunneling microscopy (STM) to study nanoscale surface topography in the 1980s, efforts have been underway to combine them with Raman spectroscopy, since SPM does not give chemical information similar to the latter. It was first demonstrated in 2000 that the SERS enhancement can be confined to a nanoscale surface region with the metal particles placed on a sharp SPM tip2. The spatial resolution of SPM and TERS is limited by the nanoscale tip dimensions rather than by far-field optics. Consequently, near-field Raman imaging with correlating morphological and chemical features in the nanometer scale can be achieved. Spatial resolution better than 20 nm has been realized by various groups, with subnanometer resolution achievable under ultrahigh vacuum
conditions.
Conventionally, electron microscopy (EM) is used along with SPM to study particle dimensions on the nanoscale. However, EM, like SPM, gives limited chemical information and may require heavy metal staining of biological samples. Alternatively, superresolution fluorescence microscopy techniques have been developed to obtain chemical information beyond the diffraction limit3,4. Such techniques still use fluorescent labels whose broadband signal does not provide detailed molecular information similar to Raman spectroscopy. A label-free method can prevent unwanted interaction between the label and the analyte, preserve the analyte’s function and avoid signal from the label itself, thus achieving a high degree of specificity with regard to chemical identity or characterization. TERS can provide this chemical information with nanoscale resolution, under label-free, ambient conditions.
Experimental design
A detailed description of TERS experimental design and applications may be found elsewhere5,6,7. Laser illumination of the tip-sample junction and signal collection can be carried out in four different configurations: bottom, top, side with microscope objectives, or a parabolic mirror with aperture to enable the laser to pass to the sample (Figure 1). While each configuration has its own merits and drawbacks, bottom access is preferred for studying transparent materials, whereas other configurations are suitable for opaque materials. The choice of laser polarization must be considered for different illumination angles; the polarization plays an important role in enhancing the electromagnetic field directly below the tip apex but also selectively excites specific vibrational transitions.
The tip is the most important part of the design as it controls spatial resolution and signal enhancement. Upon laser illumination with appropriate polarization, charge density is increased at the metallized tip apex due to the lightning rod effect. If the laser wavelength matches the surface plasmon resonance, an enhanced electromagnetic field is created with high spatial confinement. As a result, the near-field Raman signal intensity is enhanced compared to the far-field background, with the enhancement factor reaching up to six orders of magnitude. Chemical and mechanical interactions between tip and analyte can provide further effects, which is attributed to changes in the electronic structure of the analyte. Such interactions can change spectral features such as peak position and width, which may be avoided by regulating the tip-sample distance accordingly.
The excitation wavelength can be selected to match the optical properties of the noble metal on the tip. Silver and gold coatings are commonly used with 532 and 633/785 nm, respectively. Apart from typical SPM substrates like glass and mica, atomically flat silver and gold nanoplates can be used as TERS substrates, providing additional enhancement due to the “gap mode” effect8.
Applications of TERS in biology
Initial TERS studies focused on pure components like nucleobases and amino acids. TER spectra have been reported for all normal nucleobases, showing characteristic peaks with enhanced signal compared to the far-field spectrum. The plasmonic enhancement of Raman signal allows label-free detection of nucleobases in picomolar quantities9. TERS studies also showed that distinct signatures of different nucleobases can be detected in RNA and DNA, thereby opening up opportunities for a direct, label-free method for nucleic acid sequencing (Figure 2); in principle, this can also be used to detect chemical modification in nucleic acids, including epigenetic changes.
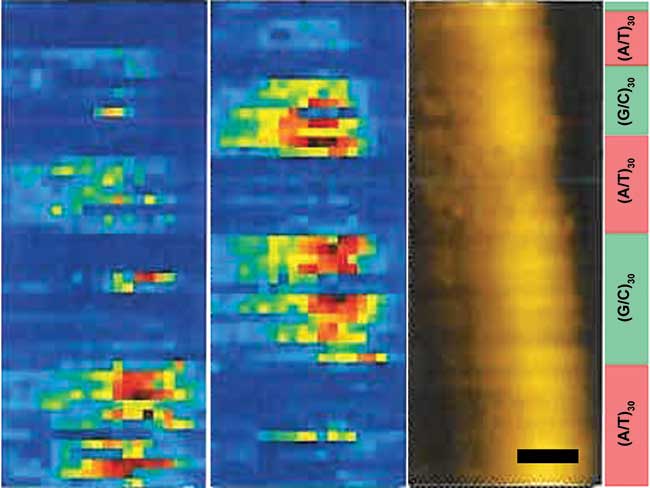
Figure 2. TERS map of A/T rich regions (left), G/C rich regions (middle) and AFM topography (right) of dsDNA (scale bar: 5 nm). The DNA sequence shown on the right was engineered from custom oligonucleotides. Adapted with permission from N.J. Kolodziejski, et al. (2014). Tip-enhanced Raman spectroscopy: Focusing in on a direct sequencing method for oxidized DNA, Biomedical Optics, Miami, Fla. Optical Society of America. Copyright 2014 Optical Society of America.
TERS has been used for surface characterization of amyloid fibrils, which are macromolecular protein assemblies implicated in neurodegenerative diseases, elucidating the surface compositions of amino acids and secondary structural conformations. This showcases the potential of TERS to study fibril surface structure, which is poorly understood in general compared to overall fibril structure. More recently, TERS studies were extended toward discriminating polymorphs of insulin fibrils also based on surface composition10. Notably, different fibril polymorphs have been associated with different cytotoxicity levels.
A significant recent development is simultaneous topographic and TERS imaging, which can correlate topographic features with a near-field Raman spectrum from every pixel of the image. The full potential of TERS imaging for studying biomaterials is yet to be realized. However, this potential can be seen in a TERS image collected from Aβ(16-22) peptide nanotapes (Figure 3), which is based on the 1004 cm-1 aromatic marker band intensity11. Besides image correlation, nanotapes not located by topography could be identified from their chemical signals.
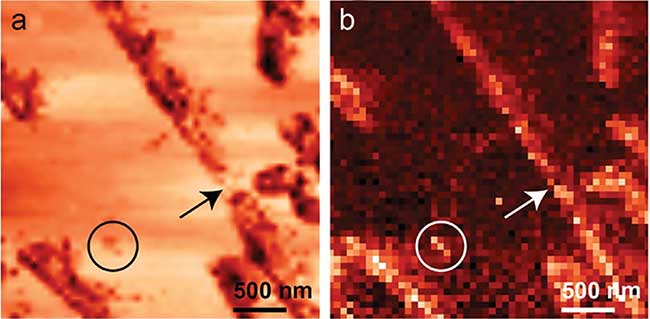
Figure 3. Simultaneously acquired (a) STM and (b) TERS (acquisition time 1 s/pixel, 2 mW incident power) images of individual Ab(16-22) peptide nanotapes with 50 × 50 pixels. The color-coded TERS image displays the peak integral value corresponding to the contribution of 1004 cm-1 aromatic marker band (brighter pixels represent higher intensity). Features weakly observed in STM image (circle and arrow) can be identified as nanotape based on TERS. Adapted with permission from M. Paulite, et al. (2013). Full spectroscopic tip-enhanced Raman imaging of single nanotapes formed from β-amyloid (1-40) peptide fragments. ACS Nano, Vol. 7, pp. 911-920. Copyright 2013 American Chemical Society.
Complex biological systems
In addition to studying multicomponent samples like nucleic acids and amyloid fibrils, TERS has been used to study the biochemical composition of other complex biological systems, such as the surface of a virus, bacterium or a human cell. For example, TER spectra have been obtained from the surface of a tobacco mosaic virus that shows specific chemical signals from viral coat proteins and RNA. More recently, TERS was used to distinguish between Varicella-zoster virus and porcine teschovirus based on differences in their surface protein and lipid composition12.
Of particular note is the detection of a membrane protein from a human erythrocyte under aqueous conditions, with both tip and analyte immersed13. This approach, which remains to be explored further, offers the advantages of studying biomaterials in their native environment and minimizing sample decomposition due to laser-induced heating and oxygen-mediated photobleaching. One interested in studying large, complex heterogeneous cell surfaces might face the problem of locating nanoscale features of interest. To address this, a targeted approach of using antibody-conjugated nanoparticles that are detectable by dark-field microscopy has been proposed14; the specific interaction between antibody and cell surface antigens can then be probed by TERS (Figure 4). This approach has enabled the detection of integrin receptors on intact cell membranes15.
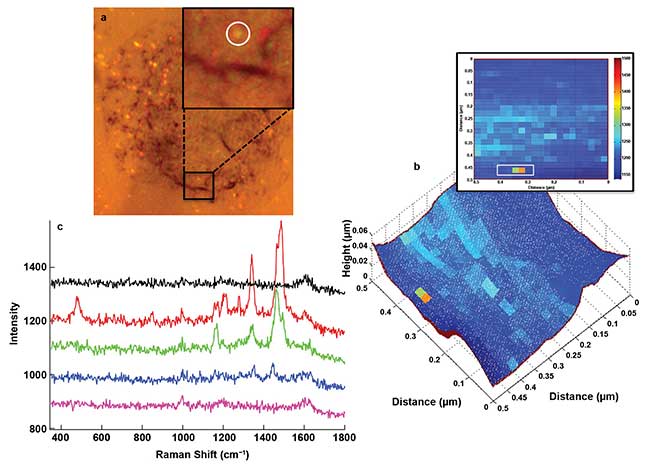
Figure 4. (a) Bright-field image of 50-nm Au nanoparticles on a cell. (Inset) Region of the surface imaged by TERS. (b) AFM topography map overlaid with TERS image built with baseline subtracted Raman intensity of 1485 cm-1 peak corresponding to Tyr residue of IgG antibody conjugated to Au nanoparticle, with 2D image of the latter shown in inset. (c) Raman spectra of IgG antibody obtained from a single Au nanoparticle corresponding to pixels within the white box (b, inset). The observed peaks were assigned to aromatic amino acids of IgG antibody, as well as the result of an IgG antibody-antigen interaction on the cell surface. Adapted with permission from K.D. Alexander and Z.D. Schultz (2012). Tip-enhanced Raman detection of antibody conjugated nanoparticles on cellular membranes. Anal Chem, Vol. 84, pp. 7408-7414. Copyright 2012 American Chemical Society.
At a ‘tip-ping’ point
Simultaneous with advances in biological applications, advances in instrument capabilities have been made. Correlated topographic and TER hyperspectral imaging capabilities, as opposed to single point measurements, are commercially available. Spatial resolution has been improved from ~50 nm in earlier studies to capabilities better than 20 nm. Tip-pressurized method and ultrahigh vacuum TERS can be used to achieve nanometer resolution.
Application of TERS has not been widespread due to challenges in preparation of robust, reliable tips. The stability of a tip and the enhancement that it provides are influenced by the interplay of many factors such as tip material, type and size of metal particles to name a few16. The lack of a universal tip preparation method has required new users to standardize a method before application. Tip-related artifacts such as carbon contamination must be avoided. The advantages of longer data acquisition from materials with low sensitivity may be limited by thermal drift during measurement. As TERS probes a smaller sample volume compared to normal Raman, sensitivity to local interactions can lead to spectral fluctuations17.
Access to tips that remain stable and provide reproducible enhancement is critical. Toward this end, commercial gold and silver TERS tips have recently been developed, which are available through companies such as Horiba Scientific and AIST-NT. Together with advances in instrumentation such as imaging18, this is expected to increase efforts toward applications. Tips providing higher contrast can enable faster acquisition and imaging, and can minimize drift. Other areas of research have emerged, such as combining TERS with UV excitation, and with pulsed lasers. Although TERS has not been a commonly used technique, due to its unique set of capabilities and recent developments in instrumentation, it is expected to provide solutions to a variety of scientific problems in the future.
Acknowledgments
The authors would like to thank Noah Kolodziejski of Radiation Monitoring Devices for valuable contributions.
Meet the authors
Maruda Shanmugasundaram is an AFM-Raman applications scientist with Horiba Instruments Inc. in Edison, N.J.; e-mail: [email protected]. Fran Adar is a Raman principal scientist with Horiba Instruments Inc. in Edison, N.J.; e-mail: [email protected].
References
1. F. Adar and M. Shanmugasundaram (April 2016). SERS spectroscopy solves outstanding problems in the biological sciences. BioPhotonics, pp. 37-40.
2. G. Sharma, et al. (July 2015). Tip-enhanced Raman scattering — Targeting structure-specific characterization for biomedical samples. Adv Drug Deliv Rev, Vol. 89, pp. 42-56.
3. L. Langelüddecke, et al. (2014). Infrared and Raman Spectroscopic Imaging. Weinheim, Germany: Wiley-VCH, p. 484.
4. N. Ji, et al. (2008). Advances in the speed and resolution of light microscopy. Curr Opin Neurobiol, Vol. 18, pp. 605-616.
5. G. Sharma, et al., pp. 42-56.
6. L. Langelüddecke, et al., pp. 479-505.
7. N. Kumar, et al. (2015). Tip-enhanced Raman spectroscopy: Principles and applications. EPJ Techniques Instrum, Vol. 2, p. 9.
8. N.J. Halas, et al. (2011). Plasmons in strongly coupled metallic nanostructures. Chem Rev, Vol. 111, pp. 3913-3961.
9. K. F. Domke, et al. (2007). Tip-enhanced Raman spectra of picomole quantities of DNA nucleobases at Au(111). J Am Chem Soc, Vol. 129, pp. 6708-6709.
10. D. Kurouski, et al. (2014). Surface characterization of insulin protofilaments and fibril polymorphs using tip-enhanced Raman spectroscopy (TERS). Biophys J, Vol. 106, pp. 263-271.
11. M. Paulite, et al. (2013). Full spectroscopic tip-enhanced Raman imaging of single nanotapes formed from β-amyloid(1-40) peptide fragments. ACS Nano, Vol. 7, pp. 911-920.
12. K. Olschewski, et al. (2015). A manual and an automatic TERS based virus discrimination. Nanoscale, Vol. 7, pp. 4545-4552.
13. B-S. Yeo, et al. (2009). Tip-enhanced Raman spectroscopy — Its status, challenges and future directions. Chem Phys Lett, Vol. 472, pp. 1-13.
14. K.D. Alexander and Z.D. Schultz (2012). Tip-enhanced Raman detection of antibody conjugated nanoparticles on cellular membranes. Anal Chem, Vol. 84, pp. 7408-7414.
15. H. Wang and Z.D. Schultz (2014). TERS detection of αvβ3 integrins in intact cell membranes. Chem Phys Chem, Vol. 15, pp. 3944-3949.
16. B-S. Yeo, et al. pp. 1-13.
17. L. Langelüddecke, et al., pp. 479-505.
18. A. Krayev, et al. (2014). High-speed TERS imaging: The latest achievements in nano-Raman spectroscopy. Spectroscopy, Vol. 29, pp. 40-49.
See “SERS Spectroscopy Solves Outstanding Problems in the Biological Sciences,” also by Fran Adar and Maruda Shanmugasundaram, in the April 2016 issue of BioPhotonics.