3-D techniques advance a range of biological research studies.
Conventional light microscopy is generally constrained by the diffraction limit, the fundamental maximum resolution of an optical imaging system resulting from the diffraction of light. In more recent years, however, researchers have developed a host of techniques – broadly known as superresolution imaging techniques – that enable them to overcome the diffraction limit.
The implications of this for biological research were considerable – so considerable that Nature Methods named superresolution imaging the 2008 Method of the Year. “As electron microscopy did in the past, superresolution microscopy, or nanoscopy, provides the ability to see details of cellular and even macromolecular structure that were not possible to see before,” the editors wrote when explaining their selection.
“Notably, however, nanoscopy is compatible with live cells and has the capability for multiplex labeling with high molecular specificity.”
Superresolution techniques have already made important contributions in biological research. Now, with the introduction of 3-D capabilities, they have the potential to make an even greater impact.
One example is the technique known as superresolution photoactivated localization microscopy, or PALM. Here, photoswitchable fluorescent molecules are sparsely and stochastically activated so that only one in a field of many will be in the “on” state at a time. This makes it possible to distinguish the emissions from those of neighboring molecules, which overcomes the diffraction limit.
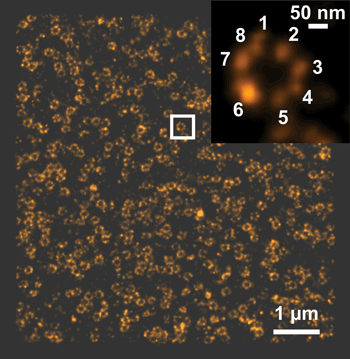
Zeiss has introduced a module that enables photoactivated localization microscopy, or PALM, in 3-D and with a lateral resolution of 20 to 30 nm and an axial resolution of 50 to 80 nm. Shown is a PALM nuclear pore image of epithelial kidney cells. Courtesy of Anna Löschberger and Markus Sauer, University of Würzburg, Germany.
At the 2013 Society for Neuroscience annual meeting in November, Zeiss introduced a module called ELYRA P.1 that enables PALM in 3-D, with a lateral resolution of 20 to 30 nm and an axial resolution of 50 to 80 nm.
The module combines two superresolution techniques – PALM and structured illumination microscopy (SIM) – in a single system, giving the user the option of choosing the technique most appropriate for his or her sample.
“In SIM, we designed the system to give the maximum resolution for each color and the optics to give a large field of view to capture large areas of interest in one go,” said Dr. Klaus Weisshart, product manager in the Biosciences Div. at Carl Zeiss Microscopy GmbH in Jena, Germany. “In PALM, we designed the system flexible enough to deal with most of the switchable fluorescent proteins and organic dyes on the market, and we improved algorithms that allowed higher accuracies even with more problematic samples.”
A combination of the two techniques also can be beneficial, he added, as it can provide structural context to localized molecules.
PALM isn’t the only superresolution imaging technique getting the 3-D treatment. In a recently published Journal of Biophotonics paper, for example, a team of researchers at Imperial College London describe 3-D stimulated emission depletion (STED) microscopy with programmable aberration correction.
STED microscopy works by suppressing, or depleting, the fluorescence signal in a region of interest – using a second beam overlapping the fluorescence excitation beam – and effectively narrowing the point-spread function to below the diffraction limit by having a minimum intensity at the center of the second beam.
The setup described in the Journal of Biophotonics paper achieves a 3-D depletion point-spread function by incorporating a liquid crystal spatial light modulator (SLM) device to produce the desired depletion beam profiles. “We believe this presents a very practical and convenient approach to 3-D-STED,” said Hugo Sinclair, the study’s corresponding author. “The use of an SLM also allows us to correct for optical aberrations in the instrument and sample.”
Sinclair and colleagues initially developed the microscope to enable superresolved imaging of biological samples, focusing in particular on the immunological synapse between interacting cells such as between a natural killer cell and its target cell – here, he said, the synapse is typically aligned perpendicular to the coverslip, so improved resolution in both the lateral and axial directions is desirable. But they anticipate that the technique could be applied to a wide range of additional biological samples, including interacting cells and 3-D cell cultures.
They continue to refine the technique with this in mind. For example, they are further developing the adaptive optics schemes used to correct for sample aberrations. “More advanced methods may be required in the presence of higher-order aberrations stemming from complex refractive index structure in samples,” Sinclair said. They also are working to incorporate fluorescence lifetime imaging to enable multilabel 3-D STED microscopy.
The Gathering STORM in Biological Research
Dr. Michael Börsch and colleagues at Jena University Hospital and Friedrich Schiller University in Jena, Germany, have been studying FoF1-ATP synthase for the past 15 years, using single-molecule Förster resonance energy transfer (FRET) to study the membrane enzyme in vitro. But when it came time to measure the static/dynamic distribution of FoF1-ATP synthase in E. coli cells so they could measure the rotary motors of the enzyme in vivo, they turned to 3-D superresolution microscopy.
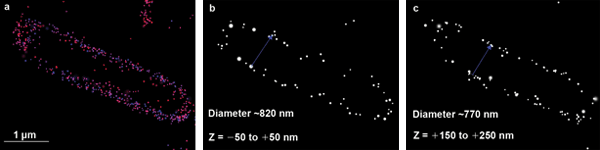
After exploring several possibilities, the researchers went with a structured illumination microscopy (SIM)/stochastic optical reconstruction microscopy (STORM) instrument from Nikon. “It was kind of a big scandal here in Jena, where everyone has a Zeiss laser-scanning microscope,” Börsch said, “but the Nikon is providing us with important information.”
The researchers reported their findings at the BiOS/Photonics West meeting in February. Using 3-D STORM and PALM as well as SIM, they probed the spatial distribution and diffusion properties of bacterial FoF1-ATP synthases in living E. coli cells. This enabled them to determine the optimal label strategy for their single-molecule FRET studies of the rotary motors of FoF1-ATP synthase in vivo.
Shown here are images of 3-D localized FoF1-ATP synthases in a living E. coli cell. The Nikon SIM/STORM microscope used to obtain the images incorporates the Agilent 4-laser-box, Nikon’s Perfect Focus System (autofocus) and an Andor EMCCD IXON Ultra camera. Images courtesy of Anja Renz/Michael Börsch, Jena University Hospital.