One technology that could make personalized medicine possible is quantitative real-time PCR, but for personalized medicine to become the norm in clinical settings, the technique must be improved.
David L. Shenkenberg, Features Editor
Medicine in the near future will be based more on each patient’s unique genetic code – a phenomenon often referred to as personalized medicine.1–3 Certain drugs already target proteins produced in patients or replace proteins not produced as a result of genetics. These proteins and other molecular indicators of disease have been called biomarkers.
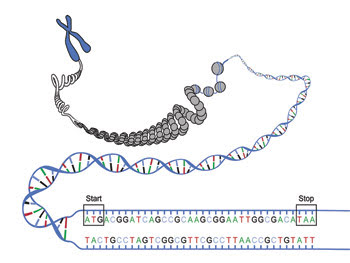
Some experts say that genetic technology is ready for personalized medicine, while others say it needs improvement; even those who say that the technology is there note that interpreting the data is problematic.
According to assistant professor Dan Ehrlich at the Whitehead Institute for Biomedical Research in Cambridge, Mass., “The game is a lot more than biomarkers – it is understanding the regulatory networks.” He and his colleagues have been designing machines that rapidly analyze genetic material.
The knowledge gained from sequencing the entire genome of individual patients could spur medical research and development toward personalized medicine. The first human genome was sequenced in 2001. Although it was a milestone of scientific and human achievement, it did not account for variation among humans. Many projects have aimed to do that.4
By mid-2007, the International HapMap Project had catalogued more than 3 million of an estimated 15 million single-nucleotide polymorphisms – places along the genome where just one of the adenine, guanine, thymine or cytosine bases is different. “Eventually, specific information on single-nucleotide polymorphisms will help in choosing between medications,” Ehrlich said. “This will fuel personal sequencing, but the database isn’t there yet.”
In Cambridge, UK, the Wellcome Trust Sanger Institute has directed efforts toward identifying genetic changes that cause disease. On July 1, 2008, the Wellcome Trust announced that its database of genetic information had reached 1 terabase, or 1 trillion bases, and that the institute produces as much sequence every two minutes as was generated in the first five years of all of the international sequence databases.5
Meanwhile, on Sept. 18, 2008, researchers from the McLaughlin-Rotman Centre for Global Health in Toronto announced that Mexico, India, Thailand and South Africa were setting an example for other developing countries in the domain of personalized medicine.6 These countries have embraced the technology because it could help poor patients avoid unnecessary medical care. “Developing countries cannot rely on the altruism of Western economic interests to address specific health needs of their populations,” said professor Abdallah Daar of the Centre.
However, problems exist with these grand notions of personalized medicine. One concern has been that employers could use genetic information to discriminate against workers and that insurance companies could use the data to disqualify patients or to charge higher premiums. The Genetic Information Nondiscrimination Act is an example of legislation designed to prevent employer and insurance company discrimination.7 It became US federal law on May 21, 2008.
The second major hindrance is cost. The original human genome project cost about $3 billion – well beyond the range of the average consumer and certainly a barrier to research as well. The National Human Genome Research Institute has set a goal of $1000 per genome. In August of this year, it announced more than $20 million in grants funding toward this goal.8–10
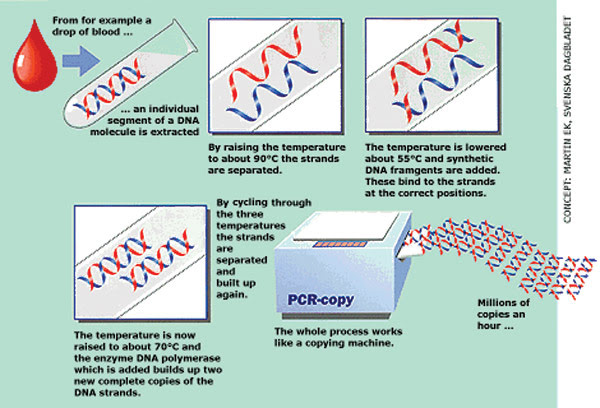
The polymerase chain reaction (PCR) has revolutionized biology, medicine and forensics. It mimics DNA replication as it occurs in cells. Short DNA sequences called primers bind to the DNA template, and the DNA goes through heat-up and cool-down temperature cycles. A DNA polymerase from Thermus aquaticus (Taq) then makes more DNA with each cooldown.
“Although Roche, Applied Biosystems and Illumina have recently come out with new systems for fast sequencing of whole genomes, these techniques are still far from the goal of $1000 per genome,” according to Ward De Spiegelaere of Ghent University in Belgium. He regularly performs quantitative real-time polymerase chain reaction (PCR), often abbreviated as qPCR, and he and his colleagues describe its optimization in a recent paper.11
Quantitative PCR is one technology that could make the genetic revolution possible. Ehrlich pointed out that a person’s health changes over the years, but the genome stays basically the same. “Expression analysis from qPCR will be far more informative than (genomic) sequencing,” he said.
Willard Freeman, who has seen a million qPCR reactions a year for the past 10 years, weighed in with his opinion. An assistant professor of pharmacology, he is director of the Functional Genomics Core Facility at Penn State College of Medicine in Hershey, Pa., and recently reviewed qPCR.12
Freeman said, “In my personal opinion, the public focus on disease susceptibility is probably misguided (because) most genetic factors contribute only a small amount to the likelihood of developing a disease, and environmental factors … play a huge role.” He believes that pharmacogenetics – the science of how the body absorbs, metabolizes, distributes and excretes a drug – may have more immediate impact on medical care.
State of the art
Freeman emphasized that real-time qPCR has matured to the point that his group and many others no longer worry about optimizing it. “Instead, we do biology,” he said.
Of the few types of fluorescent probes available for qPCR, only two are used by most researchers: TaqMan and SYBR Green. Freeman said that about 99 percent of the qPCR reactions he oversees have used TaqMan.
He criticized SYBR Green because it is only a dye that can bind to any DNA, including parts of DNA made by accident during PCR, called nonspecific amplification products. These products can be formed for several reasons; sample impurities are one factor.
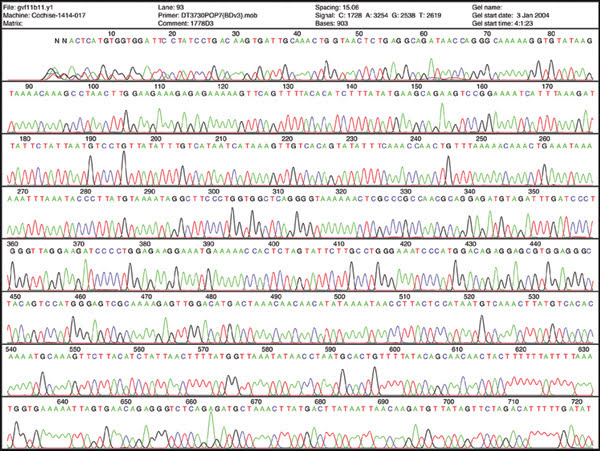
Sequencing DNA may help scientists understand disease, but some experts say that for advancing personalized medicine, quantifying messenger RNA will prove more useful. This image shows sequencing data.
In contrast, TaqMan includes a dye and a molecule called a quencher that prevents the dye from fluorescing. The dye and quencher are on a DNA sequence that matches the desired DNA sequence in the sample. Because they have to match and because the fluorescence is quenched until PCR is completed, the probes are very specific and sensitive to the DNA that scientists want to measure.
However, as De Spiegelaere points out, SYBR Green costs a lot less than TaqMan. “With TaqMan probes, for every gene you have to analyze, you have to make a new probe, and it’s really expensive.”
For optimizing PCR, De Spiegelaere and colleagues design their primers and determine the optimal primer concentration and temperature through a combination of empirical testing and software. They use the freeware programs Primer3, which designs the primer, and Mfold, which can prevent the instances when each DNA molecule will fold up and bind on itself. When this happens, the Taq polymerase may have trouble reaching the DNA, and the reaction thus may not proceed as desired.
For statistical analysis, they use two programs developed at Ghent University Hospital by Jo Vandesompele and others called qBase and geNorm. In every assay, De Spiegelaere’s team normalizes the data using several housekeeping genes, or genes that are always expressed as messenger RNA in cells. For better results, they try to use at least three housekeeping genes, analyzing the differences with qBase. Formerly available on-line as freeware, qBase and geNorm are now available in a bundled commercial software package called Biogazelle.
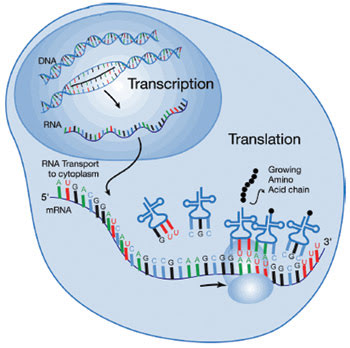
A basic concept in molecular biology is that RNA is a message that is transcribed from DNA; the message – the messenger RNA – travels to ribosomes that translate the RNA message into proteins. Although critics have derided this concept as the “central dogma of molecular biology” because the situation has proved more complicated, the idea remains an important fundamental.
Freeman noted that validated primer probe sets are commercially available, so designing primers is no longer necessary. For statistical analysis, his group employs the 2−ΔΔCT method, and he uses the software that comes with the instruments.13
Hell, thy name is RNase
Reverse transcription PCR is used for quantifying messenger RNA. Because RNA is an unstable molecule that breaks down rapidly, an enzyme from a virus is used to convert the RNA to DNA. After the reverse transcription converts the RNA to DNA, the process is identical to qPCR on DNA.
Before the RNA can be converted, it must be extracted from animal or human tissue. However, RNase enzymes that destroy messenger RNA are everywhere in tissue and even in the laboratory, unless a sterile environment is maintained.
De Spiegelaere, who quantifies RNA with reverse transcription PCR on a regular basis, said, “The processing has to be quick, and you really need to take care that you’re working with a sterile and RNase-free product.” During the RNA extraction and purification, scientists must rush around so that it does not degrade. Although some products claim to preserve RNA, the literature has not been clear on how well they work, “so we just work fast.”
“Rather than being a specifically molecular biology technique, it might be a sample handling improvement (that destroys RNases),” Freeman said, noting that some groups have used anesthetized mice and then a focused beam of microwaves on the brain to simultaneously euthanize the mice and inactivate the RNases in the brain tissue.
Other work has focused on trying to extract RNA from formalin-fixed paraffin-embedded tissue. Pathology labs typically preserve tissue this way, and it gets rid of the RNases but makes reaching the RNA difficult.
A product often used to keep lab benchtops free of RNases is called RNase AWAY. “We use it because it’s a good detergent,” De Spiegelaere said, adding that, to kill RNases, they bake their glassware for two hours at 180°.
Some of the latest reverse transcription qPCR research has centered on analyzing microRNAs – RNA molecules that are only about 20 bases long.14 Although some roles have been found for these tiny pieces of RNA, what microRNAs do remains an active area of investigation. Scientists have had difficulty quantifying them because they are so tiny and because the primers and Taq polymerase just won’t fit. Or, as Freeman puts it, “There’s not enough real estate.” However, scientists have found ways around this, such as adding bases onto the microRNA being analyzed.
Fast and cheap
Recently, researchers have added to their probes diverse fluorophores with various excitation and emission wavelength combinations to do “multiplex” PCR – the amplification of multiple genes in the same test tube. Several groups have used microfluidic technology and high-resolution imaging to analyze multiple targets quickly.
Freeman said that the companies BioTrove Corp. of Woburn, Mass., and Fluidigm Corp. of San Francisco have been leaders in developing this technology. The small reaction volumes that flow through such microfluidic chips have been enabled by predeposited genetic material in microfluidic chips or by special loading instruments.
Because the reagents used in the technique are costly, the small volumes help lower the expense of real-time qPCR. The basic thermal cyclers have decreased in price, but the instruments and robotics for high-throughput real-time qPCR still represent a significant investment. Freeman said that instruments cost $100,000 or more but that many people can use the same instrument.
Ehrlich and colleagues describe in a recent paper an affordable way to perform qPCR.15 “We save a lot of money by economizing on expensive reagents and using much less sample,” he said. “Instrument costs are usually not much of a concern compared to running costs.”
As a first step, they reverse transcribed messenger RNA to DNA. They save money in the next step by avoiding using a thermal cycler specifically designed for qPCR. Instead, they use a standard PCR thermal cycler and amplify several genes in the same well using fluorescently labeled primers – probes that are similar to TaqMan but less costly, Ehrlich said.
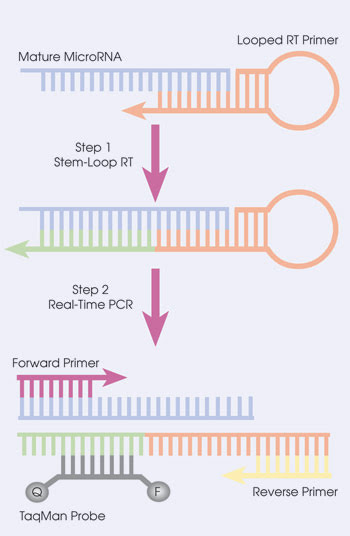
TaqMan probes have a fluorescent dye and quencher. When Taq polymerase comes along, it releases the fluorophore, and you can see the fluorescence.
After two rounds of PCR, the researchers inject the sample into a microelectrophoresis device, which is basically a microfluidic chip with places to put electrodes. The electric field separates the DNA. They then use a laser to excite the fluorescence, which is relayed by dichroic mirrors to four photomultiplier tubes, where the fluorescence is analyzed. “We need the very highest signal-to-noise ratio and need the (photomultiplier tube).”
He projects that at least 20 and as many as 50 genes can be amplified in the same test tube in a standard PCR thermal cycler. It therefore would use at least 5 percent of the enzymes and RNA of a standard real-time PCR reaction.
The researchers state that an automated microfluidic or capillary array sequencer could be modified according to the scheme they present in the paper. Using this scheme with automated technology could enable analysis of about 384 samples per hour, assuming a 384-lane microfluidic instrument. A total of 20 to 50 genes in each sample at 384 samples per hour equals 7680 to 19,200 genes per hour.
Although quantitative real-time PCR and its variants have become mature technologies for PhD-level technicians in academic laboratories, the technology is too complex and expensive to serve the personalized medicine revolution. Using heat to kill RNases, and microfluidics to hasten the process and lower reaction volumes, along with other innovations, will lower cost and improve ease of use.
References
1. B. Healy (July 28, 2008). Med school’s hot new field: Personalized medicine. US News & World Report.
2. R. Mullin (Feb. 11, 2008). Personalized medicine. Chemical & Engineering News, pp. 17-27.
3. C. Arnst et al (Oct. 18, 2004). The waning of the blockbuster drug. BusinessWeek.
4. E. Pennisi (Dec. 21, 2007). Breakthrough of the year: Human genetic variation. Science, pp. 1842-1843.
5. D. Powell (July 1, 2008). 15 human genomes each week. www.sanger.ac.uk.
6. B. Séguin et al (October 2008). Human genomic variation initiatives in emerging economies and developing countries. Nature Reviews Genetics, S3-S4.
7. www.genome.gov/24519851
8. E.R. Mardis (July 27, 2006). Anticipating the $1000 genome. Genome Biology.
9. R.F. Service (March 17, 2006). The race for the $1000 genome. Science, pp. 1544-1546.
10. NIH promises funds for cheaper DNA sequencing. Nature. August 2008.
11. W. De Spiegelaere et al (Nov. 1, 2008). Elimination of amplification artifacts in real- time reverse transcription PCR using laser capture microdissected samples. Analytical Biochemistry, pp. 72-74.
12. H.D. VanGuilder et al (April 2008). Twenty-five years of quantitative PCR for gene expression. BioTechniques, S619-S626.
13. K.M. Livak and T.D. Schmittgen (December 2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(2Delta Delta C(T)) method. Methods, pp. 402-408.
14. R.A. Ach et al (Sept. 11, 2008). Measuring microRNAs: Comparisons of microarray and quantitative PCR measurements, and of different total RNA prep methods. BMC Biotechnology.
15. Jörn Ueberfeld et al (Oct. 1, 2008). Reaction-mapped quantitative multiplexed polymerase chain reaction on a microfluidic device. Analytical Chemistry, pp. 7430-7436.
16. www.23andme.com
Central Dogma
Freshman biology textbooks teach that RNA molecules are made using DNA as a template and that those messenger RNA molecules then move to large protein complexes called ribosomes that can make new proteins using the RNA as an instruction manual. The process is compared with a message in words. The messenger RNA is said to be transcribed from the DNA template, and the ribosomes are said to translate the message from the messenger RNA into proteins. Although the DNA to RNA to protein concept remains true from a fundamental standpoint, it has been derided as “the central dogma of molecular biology” because the situation is more complicated than that.
Parts of messenger RNA molecules may be cut out before they reach ribosomes, and sometimes messenger RNA can be prevented from reaching the ribosomes at all. Therefore, scientists cannot measure protein levels while ignoring messenger RNA levels. The complexity of the situation is one reason that measuring messenger RNA levels at various time periods is important.
The polymerase chain reaction (PCR) has revolutionized biology, medicine and forensics. It mimics DNA replication as it occurs in cells. Short DNA sequences called primers bind to the DNA template, and the DNA goes through heat-up and cool-down temperature cycles. A DNA polymerase from Thermus aquaticus (Taq) then makes more DNA with each cooldown.
A PCR Primer
The 1993 Nobel Prize for chemistry recognized Kary Mullis for the invention of PCR because the technique has revolutionized medicine, forensics, biology and biochemistry. Since then, variations of PCR have emerged that have broadened its application – for example, real-time qPCR for measuring DNA and reverse transcription real-time qPCR for quantifying RNA.
Now that the human genome has been sequenced, scientists are thinking big when it comes to DNA and RNA. Demand has increased for faster, less expensive PCR that can quantify multiple RNA and DNA molecules at once.
Essentially, PCR mimics DNA replication as it occurs in the cell. Short sequences of DNA called primers are mixed in test tubes along with the DNA sequence of interest and DNA polymerase, an enzyme that copies the DNA sequence of interest and thus produces more of it. The DNA polymerase comes from a heat-tolerant organism, usually from Thermus aquaticus (Taq). The mixture in the test tubes is kept at a certain temperature at which the polymerase and primers can bind to the template DNA; then the mixture is heated so that the primers and polymerase separate from the strand. Next the temperature is lowered to allow the polymerase and primers to bind again for another round. The process is repeated many times to make a lot of DNA. In quantitative PCR, fluorescent probes are used to quantify exactly how much DNA is there to begin with.
Personalized Genetics to Go
On the lighter side, in 2007, Anne Wojcicki, wife of Google co-founder Sergey Brin, launched 23andMe, named after the 23 chromosomes in human cells.16 The 23andMe process works like this: The company mails the client a kit with a test tube, and the client deposits his saliva in the test tube and mails it back to the company, which analyzes the saliva with an Illumina microarray system that uses a proprietary set of genes. The company’s lab personnel scan for genetic loci that encode more than 80 traits, from eye and hair color to serious diseases like rheumatoid arthritis. On Sept. 9, 2008, the company announced that it had partnered with Ancestry.com and had lowered its price to $399.