Improved spatial resolution enables the study of biological systems on the nanometer scale.
Lauren I. Rugani, Contributing Editor
Optical trapping has been utilized in endeavors as varied as microfabrication,
particle sorting and the study of nonequilibrium thermodynamics. The technique,
also known as optical tweezing, is particularly beneficial for studies on the cellular
level because it can isolate a system of interest from its environment, and it provides
a noninvasive, nondamaging method for analysis.
Continuous innovation and instrument refinement
have overcome many challenges in the field of optical trapping, allowing investigators
to study the smallest biological systems that operate within a cell. Observing the
chemical, structural and physical properties of subcellular entities eventually
will help answer some of the most fundamental questions about living beings.
Characterizing chromosomes
One benefit of optical trapping is its ability
to hold objects away from a surface. Suspending an object in an optical trap enables
more accurate analysis because it reduces the amount of surface-induced background
noise, or morphological effects. This is especially useful when the object of interest
is easily affected by interference, as are human chromosomes.
Chromosomes typically are identified
by stain-based techniques that can alter the molecular state of the chromosome,
thereby compromising its accurate analysis. As reported in the June 12 issue of
Optics Express, Yong-Qing Li and a team of researchers at East Carolina University
in Greenville, N.C., applied laser tweezers and Raman spectroscopy to trap individual
human chromosomes and to identify them by their spectral patterns.
The capability of Raman spectroscopy
to reveal molecular structure will enable the development of techniques for the
detection of chromosomal aberrations that cannot be detected by staining. “Our
technique will also allow for rapid, automated screening and diagnosis for cancers
caused by or related to chromosomal abnormalities and changes in activation status,”
said Thomas J. McConnell, a biology professor at the university.
Because chromosomes are only about
3 μm in size, it is highly advantageous to isolate them from other parts of
the cell prior to collection of Raman spectra. To accomplish this, the researchers
engineered a slide with three wells — the first larger than the other two
— connected by a series of canals. They used the beam from a 785-nm frequency-stabilized
semiconductor laser made by Sacher Lasertechnik GmbH of Marburg, Germany, directed
through a Nikon inverted microscope, to trap and maneuver the chromosomes away
from debris.
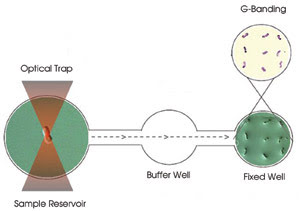
Figure 1. On a three-well slide, chromosomes are trapped in the sample
reservoir for Raman spectroscopy, maneuvered through the buffer well to avoid debris
and, finally, adhered in the fixed well for G-banding.
The process began with the trapping
of individual chromosomes in the first well. There, the same laser was used at 50
mW for Raman spectroscopy. The average Raman spectra over two 30-s readings were
collected and recorded by a Horiba Jobin Yvon spectrometer equipped with a liquid-nitrogen-cooled
CCD detector. To avoid attracting debris, the laser operated at 10 mW to maneuver
each trapped chromosome through the middle well and into the third well, where the
chromosomes adhered to the bottom of the slide (Figure 1).
After recording the position of each
chromosome in the third well, the investigators employed G-banding to identify them.
This staining technique enabled the scientists to match each chromosome with the
proper spectra (Figure 2).
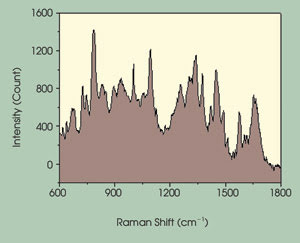
Figure 2. The Raman spectra of an individual chromosome are related to positive identification
by G-banding. The variance among spectra allows for discriminate identification
of individual chromosomes.
They compared the recorded spectra
using generalized discrimination analysis and found that the spectra could be used
to identify individual chromosomes.
“The technique could be improved
to sort the same number of chromosomes from a complex chromosome solution for more
precise analysis,” Li said. It also could potentially be used to detect differences
between one person’s chromosomes and another’s.
“Laser tweezers and Raman spectroscopy
provides a unique method to analyze biological systems, including changes in biological
systems that are due to changes in DNA and/or protein interactions,” McConnell
said.
Single-molecule studies
The team will study chromosomal breakpoints related
to leukemia and begin analyzing cancer cells to detect changes in the nuclei compared
with normal cells. It also aims to generate a library of Raman spectra for each
chromosome that will serve as a resource for research communities worldwide.
Typical single-molecule studies involving
optical trapping feature a polymer with one end attached to a fixed surface and
the other end attached to a trapped microsphere. Force applied by the optical trap
on the microsphere is used to monitor the dynamics of the polymer. In the May 23
issue of PNAS, Nicholas R. Guydosh and Steven M. Block of Stanford University
in California reported the use of this technique to measure the single-molecule
kinetics of kinesin in the presence of substrate analogues.
Conventional kinesin transports intracellular
cargo along microtubules in a hand-over-hand fashion as the two heads alternate
leading and trailing positions. The mechanism responsible for this movement remains
uncertain, however, so the researchers sought to explain how the two heads coordinate
and when the steps occur in relation to various biochemical events.
Because kinesin is small, they monitored
the molecule’s stepping patterns by trapping and following a 0.5-μm polystyrene
bead attached to one end of it. They positioned the other end of the kinesin on
a surface-bound microtubule along which it could bind and move (Figure 3).
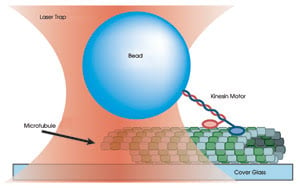
Figure 3. A 0.5-μm
polystyrene bead is held in an optical trap and bound to one end of a kinesin molecule.
The position of the bead is monitored as the two heads of the kinesin motor bind
and move along a surface-bound microtubule.
Each time a kinesin head bound to the
microtubule, an optical force clamp was activated by moving the stage to maintain
the position of the bead relative to the trap center. Because a typical force clamp
does not work for small distances or over small time intervals, the scientists developed
a more stable, low-drift apparatus. Block also has explored the use of electro-optic
deflectors in place of acousto-optic deflectors to improve the force clamps.
They formed the trap on a Nikon inverted
microscope at a fixed distance of 70 nm from the bead and used a Spectra-Physics
1064-nm diode-pumped solid-state Nd:YVO4 laser to trap the bead. Scattered light
from an 830-nm single-mode diode laser from Point Source of Hamble, UK, was monitored
by a position-sensitive detector from Pacific Silicon Sensor Inc. of Westlake Village,
Calif., to measure the bead’s position.
The scientists found that, in the presence
of ATP and beryllium fluoride, pauses lasting from <1 s up to tens of seconds
halted the progression of the kinesin molecule along the microtubule. Forward motion
could continue only after the molecule took a backward step, called a terminal backstep,
lasting a few tenths of a second. In addition, backward steps lasting milliseconds
interrupted the extended pause. Termed “recurrent backsteps,” these
motions are statistically different from terminal backsteps.
The results suggest that BeFx binds
to the rear head of kinesin and halts forward motion. To regain forward movement,
the unbound front head must take an obligatory backstep. This may be a recurrent
backstep, in which the analogue remains bound during the next forward step and kinesin
returns to its pause state, or it may be a terminal backstep, in which the bound
head releases the analogue and kinesin resumes forward stepping.
“Imagine you are walking and
suddenly someone nails your rear foot to the floor,” Block said. “Somehow,
the front leg is aware that the back leg is no longer able to move.” Similarly,
there must be some kind of signal between the two kinesin heads to coordinate stepping,
the research suggests.
When the rear head is bound, kinesin
binds ATP, takes an 8-nm step backward, hydrolizes the ATP molecule and takes either
a recurrent or a terminal backstep. Because the analogue can bind only to the rear
head, it follows that the heads are not symmetric; the nucleotide affinity of either
head must depend on its position, just as the movement of each leg is dependent
on its position relative to the other leg.
The researchers concluded that the
back head coordinates the movement of the front head and that ATP binding to the
front head is required for progressive stepping.
Next, they will probe how kinesin employs
energy from ATP for displacement. They hope to describe in molecular detail how
chemical energies in the cell are transformed to mechanical energy. “It’s
truly a fundamental question about life,” Block said.
Two beads are better than one
For high-resolution single-molecule studies that
undertake monitoring of subnanometer movements within a system, trap stability and
resolution are of great importance. Surrounding noise or drift of the components
of the trap apparatus make it difficult to take small, precise measurements. In
a dual-trap setup, a second microsphere is attached to the polymer in place of the
fixed surface. Holding each sphere in a separate trap serves to decouple the system
from surface interference. In another study, Block and his colleagues at Stanford
and at the University of Wisconsin-Madison employed such a dual-trap method to
investigate transcriptional pausing by RNA polymerase molecules (Figure 4).
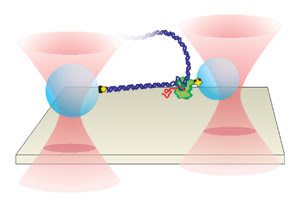
Figure 4. Separate optical
traps hold two polystyrene beads above the surface of a glass coverslip. DNA is
attached to the larger bead (left), while RNA polymerase is attached to the slightly
smaller one. The position of the smaller bead is measured to determine individual
pause sites as RNA polymerase undergoes transcription along the DNA templates.
Previous research had identified two
sequence-dependent pause classes, collectively termed “defined pauses.”
Other pause classes have been categorized by their lifetimes. Short-lifetime pauses,
also known as ubiquitous pauses, occur roughly once every 100 base pairs.
It has been difficult to establish
the relationship between ubiquitous and defined pauses, partly because previous
experiments have lacked the resolution to determine whether specific DNA sequences
initiate ubiquitous pauses.
As reported in the June 16 issue of
Cell, Block and his colleagues constructed two periodic DNA templates, each
incorporating a different pause element, and tethered them between two polystyrene
beads for the dual-trap assay. The templates were designed with repeating sequences
comprising sequence-specific pause sites. The researchers could monitor the behavior
at these sites and determine which sequences correlated with pause events.
The same optical apparatus used for
the single-trap kinesin study was employed for this experiment. However, the beam
from the trapping laser was split into two orthogonally polarized beams to trap
the two beads independently. A 700-nm bead was held in a stiff trap, while a 600-nm
bead, held in a weak trap, was illuminated by the detection laser. Position-sensitive
detectors monitored scattered light to measure the movements of the smaller bead.
The researchers identified pause sites
on both templates by combining data from many molecules and from each of the eight
repeat regions and by generating average dwell-time histograms. The data enabled
them to find a relationship between the pause positions of RNA polymerase on DNA
and the lengths and sequences of RNA transcripts.
“Every time RNA polymerase met
a sequence, it paused. We achieved near-base-pair resolution over two to three thousand
base pairs in a row,” Block noted.
The data indicated that ubiquitous
pauses are sequence-dependent, triggered by sequence signals rather than by random
events.
This confirms the idea that longer-lived
pauses — the defined pauses — arise from a common, elemental pause state
that is related to ubiquitous pausing. In this so-called two-tiered pause mechanism,
a sequence element induces an elemental pause state. Then, secondary pause mechanisms
such as hairpin formation or enzyme backtracking — examples of defined pauses
— translate the elemental pause into a long-lived one. The researchers believe
that overlapping pause-inducing and pause-stabilizing sequence elements might form
the relationship between ubiquitous and defined pauses.
In subsequent investigations, they
observed base-pair stepping at angstrom-level sensitivity to follow enzyme motion.
“We want to push resolution to the atomic level,” Block said.
They are seeking to understand gene
regulation and why different types of cells are so diverse despite having identical
DNA. They continue to study RNA polymerase and are developing methods to study and
understand transcriptional control.
Other studies support the idea that
the spatial resolution of dual-trap detection is an improvement over single-trap
designs for single-molecule studies and that the dual-trap method not only helps
isolate the system, but also lowers the effect of Brownian noise, which can hinder
the observation of small changes. Investigators led by Carlos Bustamante at the
University of California, Berkeley, presented in the June 13 issue of PNAS
both theoretical descriptions and direct measurements of the Brownian limit on spatial
resolution in optical trapping experiments.
The investigators solved the equations
of motion for two microspheres tethered by a single piece of DNA, allowing them
to predict the signal-to-noise ratio (the ratio of the observed signals from the
changes in the DNA length to the Brownian fluctuations). By making direct measurements
of the spatial resolution with their own noise-limited dual-trap optical tweezers,
they could directly test these predictions.
The two traps were formed from orthogonally
polarized beams generated by an 845-nm fiber-coupled diode laser from Lumics GmbH
in Berlin.
“We are currently upgrading this
system to a 1064-nm diode-pumped solid- state laser to allow us to form stronger
traps and exert higher forces,” explained Jeffrey R. Moffitt, a member of
the research team.
They used back focal plane interferometry
to determine the position of each microsphere relative to its corresponding trap
center. Scattered light from the microspheres was collected, separated by polarization
and imaged onto two position-sensitive photodetectors from Pacific Silicon Sensor.
In one experiment, they attached a
DNA tether to two 860-nm microspheres held in equally stiff traps. They tested the
dependence of the signal-to-noise ratio on the tension of the DNA by stretching
the tether incrementally to higher stiffness.
The measured values of the signal-to-noise
ratio agreed well with the predicted values for a wide range of stiffness, with
the difference coordinate (formed by subtracting the movements of one microsphere
from the other) providing a resolution better than that expected for a single trap.
A second experiment tested the theory
with an 800-nm microsphere in one trap and a 997-nm microsphere in the other. Again,
the scientists pulled the tether to various tensions and calculated the spatial
resolution as a function of relative stiffness between the traps.
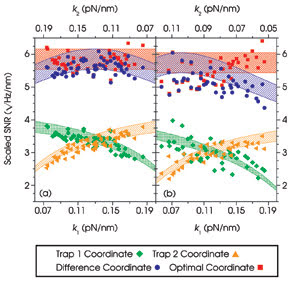
Figure 5. The measured signal-to-noise ratios (SNR) are plotted as
a function of trap stiffness (k) for two identical, tethered, 860-nm microspheres
(a) and for an 800-nm microsphere tethered to a 997-nm microsphere (b). The shaded
regions represent predicted values. Reprinted with permission from PNAS.
Once more, experiments agreed well
with the theoretical predictions; however, in this asymmetric system, the difference
coordinate was no longer the best coordinate. Instead, the researchers found that
a linear combination of the movements of the two beads, weighted appropriately to
reflect the asymmetry, produced the best resolution. The highest resolution was
achieved with the smallest possible microspheres, independent of the stiffness of
the traps (Figure 5).
Bustamante and his team found that
monitoring the movements of both microspheres enabled the detection of smaller movements
at larger bandwidths and lower DNA tensions than was possible when monitoring only
one microsphere (Figure 6). Using this technique, they showed that it is possible
to observe length changes of only one base pair (3.4 Å) at lower tensions
than previously reported (7 pN). These steps are not present when the displacement
of only one microsphere is measured.
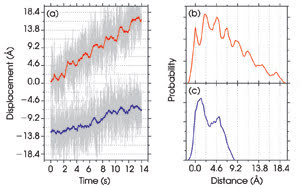
Figure 6. The displacements of the microsphere in trap 1 (blue) and
of the optimal coordinate (red) are plotted as a function of time (a). Every 2 s,
the distance between the two tethered 860-nm microspheres is increased by 3.4 Å.
Pairwise distance histograms for the optimal coordinate show peaks corresponding
to the distance between each step (b). Peaks are not present when the displacement
of one microsphere is measured (c). Reprinted with permission from PNAS.
“This will allow the direct observation
of the discrete movements of many DNA enzymes,” Moffitt said.
The improved spatial resolution of
dual-trap systems is significant for the study of biological systems that operate
on the subnanometer scale and under a limited range of forces. With this method,
the investigators plan to study the mechanisms of important molecular motors and
to explore the application of these techniques to other biological systems.
Narrowed down to size
Improving spatial resolution is one of the major
challenges in optical trapping. Until recently, typical traps could not resolve
the movements of many biological systems. Now variations of the technique have been
employed to study single-molecule dynamics, DNA manipulation by enzymes and unfolding
of individual proteins. “All of these studies could certainly benefit from
an improvement in resolution,” Moffitt remarked.
Besides improving the technique itself,
significant efforts are being made to combine optical trapping with other methods
of analysis such as fluorescence resonance energy transfer and magnetic tweezers.
As demonstrated by Li and his team,
the combination of laser tweezers with Raman spectroscopy allows researchers to
study smaller objects and obtain higher-quality results, thus gaining a fundamental
understanding of biological systems while opening doors for further research.