Optical laser traps DNA molecules and “dips” them into proteins for repair.
DNA is susceptible to continuous egregious environmental and biological hazards that can significantly damage its composition. Cellular exposure to radiation from x-rays and UV light, including free radicals and chemicals, can cause DNA to break. Fortunately, cells house many proteins aimed at repairing broken strands.
Researchers Stephen C. Kowalczykowski, a professor of microbiology and of molecular and cellular biology, and Jovencio Hilario, a postdoctoral scholar, both at the University of California, Davis, examined the human protein Rad51, designed specifically for homologous recombination – a method for mending damaged DNA and developing new combinations of DNA sequences.
To do so, they used a microfluidic flow cell, a cell-sorting system, along with fluorescence imaging and optical laser trapping to assist with and observe Rad51-DNA binding in real time.

A 3-D image depicts a single DNA-bead complex trapped by an optical laser in the center of the beam. Below it are the phases of Rad51 binding as nucleoprotein filaments extend DNA strands after multiple dipping periods over an allotted period.
Optical trapping
Optical trapping refers to manipulating microscopic objects using IR light from a focused laser beam. The beam can pick up and move nanometer- or micron-size biological particles such as cells or polystyrene beads.
Gradient forces and reflective or refractive light that acts on an object allow the beam to contain a particle within its center, creating an optical trap. The beam is focused through a microscope objective, and the narrowest point at the center attracts particles because of a high electric gradient.
The scientists used this principle with an Nd:YVO4 IR laser emitted through a fluorescence microscope objective at 1064 nm with output power of 300 to 700 mW to pick up single λ phage double-stranded DNA attached to a 1-μm streptavidin-coated microsphere at one end of the strand.
‘Dipping’ DNA
In their study, the researchers performed a process called “dipping,” where the DNA molecule is captured and moved into individual channels of a flow cell set up with various chemical cofactors. Reactions were viewed using single-molecule fluorescence microscopy.
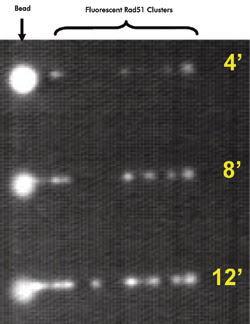
A single λ DNA molecule is shown at various times (in minutes) after a “dipping” process. Multiple nucleation events at longer time intervals revealed more nucleoprotein filament formation.
“Viewing of the complete DNA repair pathway has not been seen by single-molecule methods yet,” the scientists state. “Our methods allow us to assemble and observe protein-DNA complexes in real time, and to change reaction conditions quickly.”
Optical laser traps DNA molecules and “dips” them into proteins for repair.
The first channel – called the capture – was set up with only DNA-bead complexes labeled with YOYO-1 fluorescent dye. The second one, an observation channel, served two purposes that involved removing the YOYO-1 dye from the DNA, which may possibly have hindered Rad51 binding and later allowed for viewing Rad51-DNA assembly and disassembly.
Once the fluorescent dye was properly removed, DNA was transferred to the third, or reaction, channel, which contained Rad51 proteins labeled with carboxyfluorescein (Rad51FAM) fluorescent dye at the N-terminus. This is where dipping occurred as the DNA was incubated with Rad51 for a selected time.
The combined Rad51-DNA then was moved back into the observation channel. “Dipping could be repeated as necessary in order to observe time-dependent nucleation events or to completely coat and stretch the DNA,” Kowalczykowski said.
Repairing codes
The researchers determined that, when DNA is dipped into the third channel, two to three Rad51 monomers physically interact to create a nucleus, the critical stage of assembly. Subsequently, the nucleus grows to form a nucleoprotein filament that extends the DNA.
“The protein filament wraps around DNA and, as a result, stretches it beyond its usual length,” Kowalczykowski and Hilario noted.
Rad51 assembly took about 10 s when examined at the highest protein concentration level and nucleated along the entire DNA length. However, nucleoprotein filaments did not grow continuously; instead, they completed their process at about 2 μm in length.
With the inclusion of ATP hydrolysis in the third channel, Rad51 assembly extended DNA about 50 percent. By inhibiting ATP, hydrolysis extension was 65 percent, and the nucleoprotein filaments remained stable and extended. With the inclusion of ADP, Rad51 assembled to DNA but filament formation was not extended.

Rad51 disassembles from DNA slowly, although the process is never fully completed. The reason for partial disassembly is that the protein has a strong affinity for double-stranded DNA in the presence of ATP and ADP. As a result, other proteins must be introduced to finish the dissociation. If dissociation is incomplete, DNA synthesis and chromosome separation will not happen properly.
The examiners found the disband process to be slow – about 25 min – and incomplete because “Rad51 has a high affinity for dsDNA in the presence of ATP and ADP.”
Dissociation must be carried out for DNA synthesis – the ability to replicate – and chromosome separation to happen properly. Additional proteins, such as Rad54, may be needed to assist the process.
By manipulating and visualizing the process, the scientists determined the characteristics of Rad51 binding to DNA. Filament assembly by nucleation, growth and extension was examined, as well as disassembly, which could not be measured with ensemble experiments.
The investigators plan to include more accessory proteins in their process to discover which will enhance Rad51 assembly and which may help with disassociation.