Real-time optical re-centering of rapidly moving fluorescent targets allows functional imaging of the C. elegans nervous system.
Serge Faumont and Shawn Lockery, University of Oregon, and Gary Rondeau and John Zemek, Applied Scientific Instrumentation
It has been said that the brain works just the way you think. If only neuroscience were that simple. The basic computational element of the brain is the point of contact between two neurons, called a synapse. With 1012 neurons and 104 synapses per neuron, the human brain contains about 1016 distinct computational elements. In the face of such complexity, many neuroscientists turn to the brains of smaller, more primitive creatures. One such animal is the nematode Caenorhabditis elegans. The nervous system of this microscopic, transparent roundworm contains just 302 neurons and about 5000 synapses.
It has been possible to reconstruct the wiring diagram of the nematode brain, which is the first big step toward understanding how its tiny brain works. The next step is to define the functional role of each neuron. The ultimate way to do this is to record neuronal activity in an intact, freely moving animal while it is engaged in a variety of natural behaviors. This project is particularly satisfying in C. elegans because each of its 302 neurons is an identifiable entity that recurs in every individual of the species.
With the advent of genetically encoded optical probes of neuronal activity, neuronal activity now can be recorded in intact nematodes. But doing this in a freely behaving animal presents two key challenges. First, high-magnification microscope objectives (63× to 100×) are required to resolve the minute, tightly packed nematode neurons. These objectives have fields of view on the order of 100 μm, whereas nematodes crawl at speeds of up to 400 μm/s. Thus, the target neuron moves through the field of view in a fraction of a second. Second, even though nematodes are only about 1 mm long, just a small fraction of the worm can be seen in any frame, making it nearly impossible to correlate behavior with neuronal activity.
Imaging moving fluorescence
To address these challenges, a noncommercial partnership was formed in Eugene, Ore., between researchers at the University of Oregon and Applied Scientific Instrumentation, which offers precision microscope automation. Our goal was to create a system for real-time optical re-centering of moving fluorescent targets for functional calcium imaging.
The operating principle of the system, called PhotoTrak, is illustrated schematically in Figure 1. An inverted compound microscope is fitted with a calcium imaging camera, a quadrant photomultiplier tube (PMT) and a DC servomotor X-Y stage (MS2000-INV). A beamsplitter diverts 20 percent of the light to the PMT and, using the real-time output from the PMT — the intensity of light on each quadrant — the controller computes the stage movements required to re-center the target in the field of view. The re-centering algorithm, which involves equalizing the illumination across the PMT, is simple enough to implement with analog electronics. Thus, the stage position can be adjusted continuously with a latency of less than 25 ms, as required for re-centering targets moving at hundreds of microns per second.
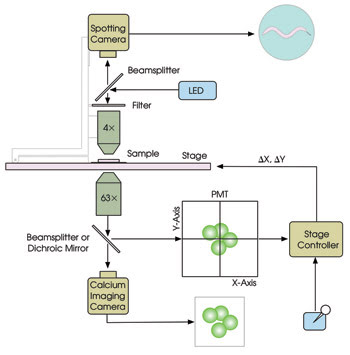
Figure 1. The main components of the PhotoTrak system for real-time optical re-centering are shown.
To record the overall behavior of the worm, wide-field images are taken from above using a spotting camera for epi-illumination at a wavelength well separated from those used for calcium imaging. The video streams from the two cameras are synchronized to provide simultaneous records of neuronal activity and behavior. The target’s X and Y coordinates, as reported in real time by the controller, are stored on disk for off-line behavioral analysis. Additionally, this information can be used to control external devices during experiments.
Optical probes provide a relatively noninvasive means of recording neuronal activity. To date, the best signal-to-noise ratios are obtained with probes for intracellular calcium concentration, which increases when neurons become more active. We prepared worms for calcium imaging by constructing an artificial gene in which the DNA sequence for the calcium probe — the so-called cameleon protein in this case — was placed under the control of a regulatory sequence (i.e., promoter) that is known to be activated only in the subset of neurons from which one wishes to record. Injecting this construct into a worm causes it to be incorporated into the worm’s DNA, thereby creating a strain of worms in which cameleon is expressed only in the desired neuronal subset. In C. elegans, cameleon expression can be restricted to exactly one neuron in certain cases. Worms expressing cameleon in small subsets of neurons are shown in Figure 2.
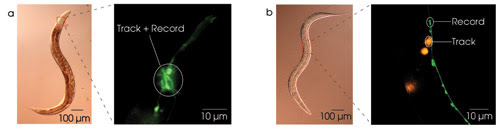
Figure 2. Large, bright fluorescent targets are required to prevent the system from tracking the worm’s naturally occurring autofluorescence. To simultaneously image calcium and behavior, the target neuron can be a member of a cluster of fluorescently labeled neurons (a), or the tracking can be done using a red proxy target in the vicinity of the target neuron (b).
One early technical challenge wefaced was that the tracking system is easily distracted by the worm’s gut, which is, unfortunately, strongly autofluorescent. One method to solve this problem is to use strains of worm in which the target neuron resides in a bright cluster of cameleon-expressing neurons (Figure 2a). Off-line image segmentation then is used to obtain the calcium signal from the target neuron.
However, to record from a single neuron in isolation — which is far dimmer than the gut — a second method must be used. Here, one creates a strain of worms that expresses cameleon in the target neuron and dsRed, a bright-red fluorescent protein, in a large non-neuronal cell close by, the latter serving as a proxy target (Figure 2b). DsRed’s excitation spectrum partially overlaps that of many calcium probes, including cameleon; thus, a single excitation source can excite both proteins. When using this approach, substituting a short-pass dichroic mirror for the beamsplitter can prevent the proxy target from appearing in the image on the calcium camera.
The two approaches are complementary. The first is simpler than the second because tracking and calcium imaging are done at a single wavelength; whereas the second approach has the advantage that all of the light from the tracking target reaches the PMT.
With the PhotoTrack system, recording neuronal activity and behavior simultaneously is relatively simple. First, the animal is prepared for recording by sandwiching it between a glass coverslip and an agar disk, which provides moisture to sustain the animal and traction for locomotion, and then it is placed on the X-Y stage. The user manually translates the worm toward the optical axis of the microscope. This is done by observing the video output of the spotting camera while using a joystick to move the stage. When the fluorescent target first becomes visible in the video output of the Ca2+ imaging camera, the user activates the tracking function via a trigger on the joystick. Subsequently, a second trigger starts the process of synchronously streaming to disk the outputs of the two cameras and the target’s X and Y coordinates.
In practice, we found that the system stably tracks targets in nematodes moving at normal crawling speeds, often for tens of minutes. When the system loses the target, it usually is because the target escaped the focal plane as a result of movement along the Z-axis. This problem can be addressed by incorporating an autofocus mechanism currently being developed at Applied Scientific Instrumentation.
Neurons for crawling
We have used the system to assign functional roles to neurons believed to be involved in regulating crawling behavior (Figure 3). Crawling in nematodes is sinuous, as it is in snakes. However, unlike snakes, nematodes can crawl in both directions, forward and reverse, and the worm modulates the relative frequency of these two modes as it navigates around obstacles and up or down chemical gradients. Thus, a key question is how this modulation is achieved at the level of individual neurons.
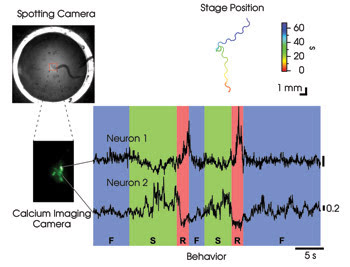
Figure 3. To assign functional roles to neurons that are active during locomotion in freely crawling nematodes, tracking was performed using the method shown in Figure 2a. Off-line image segmentation was used to isolate the calcium signals of two cells in a cluster of neurons labeled with the cameleon calcium probe. The traces in the colored graph show relative calcium concentration in arbitrary units as a function of time. Crawling behavior, scored by viewing the spotting camera video, is indicated below. F = forward; R = reverse; S = stop.
To address this question, we engineered worms that expressed the cameleon probe in a small cluster of neurons whose outputs go to distinct populations of motor neurons that directly activate the muscles for forward and reverse locomotion. We found that activity in one of these neurons (Figure 3, Neuron 1) was high only when the worm reversed, suggesting that it might be the long-hypothesized command neuron for reverse locomotion. In contrast, activity in a second neuron (Neuron 2) was high when the animal stopped and low when it reversed. This neuron may be acting like a brake on reverse crawling. Overall, the activity patterns of these neurons are consistent with a role in choosing the direction and speed of locomotion. It should now be possible to systematically survey activity patterns during behavior of almost any neuron in the nematode.
Biological data are inherently dynamic in time and space. The PhotoTrak system follows targets moving at speeds of hundreds of microns per second over regions of space that far exceed the field of view of a compound microscope. As such, it should find a wide variety of applications, including tracking high-speed objects in other organisms, such as the larvae of fruit flies and zebra fish. This system also is readily adaptable to tracking slow-speed objects over similar regions of space. Thus, it should be possible to automate time-lapse studies of gradual cellular processes at single-cell resolution. Such processes include cell migration, translocation of subcellular organelles and axonal path finding. The development of the system provides an example of how science and industry can work together to develop new tools to promote research in many fields.
Meet the authors
Serge Faumont is a postdoctoral research associate and Shawn Lockery a professor, both at the Institute of Neuroscience, University of Oregon in Eugene; e-mail: mailto:shawn@chi%20nook.uoregon.edu.
Gary Rondeau is the technical director of Applied Scientific Instrumentation in Eugene and John Zemek the executive director; e-mail: [email protected].