A simple modification allows researchers to extract more data in a single image, reducing the images needed to fully reconstruct a cell.
KATIE HEISER AND LESLIE KIMERLING, DOUBLE HELIX OPTICS LLC
Light microscopy is a powerful method used in many scientific disciplines, including the life sciences, to visualize finer details in samples. The use of fluorescence enables specific targeting and labeling of biomolecules and other chemicals. However, in light microscopy there are trade-offs between the time it takes to acquire an image, the resolution of the resulting image, and photobleaching or damage to the sample. Recent developments in the field have pushed these limitations.
A major development is superresolution imaging. This enables researchers to break the diffraction barrier and visualize subcellular structures well below the conventional 200-nm limit1. The highest resolution methods are a family of techniques known as single-molecule localization microscopy (SMLM)1. Although SMLM enables high-precision imaging of 10 to 20 nm in the lateral dimension, it typically lacks axial (Z) resolution, especially near focus.
With the addition of high-precision axial information and extended depth of field, researchers can obtain more data with more accurate information to directly measure 3D movement.
One method to extract axial information uses an astigmatic lens to distort the point spread function, enabling extraction of a limited amount of 3D information. Unfortunately, this approach has a depth capability of about 800 nm. Alternative methods developed to obtain high-precision 3D images, such as iPALM and multifocal imaging, require custom-built microscopes, optics, and software that may not be accessible to most researchers2.
An alternative technique, known as the double-helix point spread function (DH-PSF) offers a solution to this problem by enabling high-depth, high-precision 3D imaging. The DH-PSF modifies the point spread function on the microscope such that, instead of an Airy disc, the image of each point source is in the form of two well-separated lobes. These rotate around their midpoint as the emitter is moved along the axial dimension. The axial dimension of the emitter is encoded in the angle of the two lobes. The center point between them indicates the lateral position3.
With this simple modification of the microscope, researchers can extract more data with high-precision, 3D information compared to 2D SMLM (Figure 1). The increased depth of the DH-PSF captures more microtubules with high-precision axial information over a larger volume than conventional 2D SMLM. By enabling extraction of more data in a single image, the technique reduces the number of images required to fully reconstruct a cell, while improving the resolution of the image.
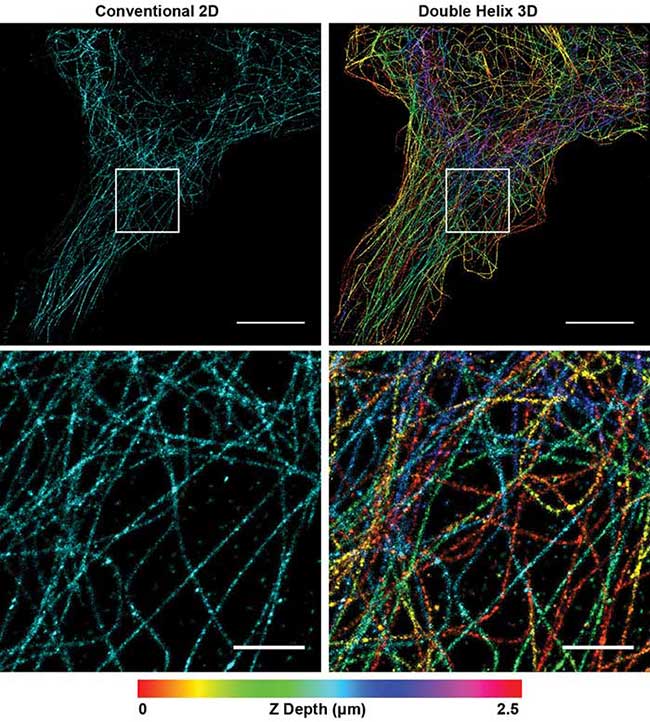
Figure 1. Superresolution image of microtubules in a Cos7 cell. Conventional 2D reconstruction (simulated, left) double helix 3D reconstruction (right). Depth encoded in color, scale at bottom. Top panel scale bars = 10 μm; bottom panel scale bars = 2 μm. Courtesy of Double Helix Optics LLC.
Using this powerful method, Jain and colleagues were able to describe core structures in RNA stress granules for the first time4. Constituted of RNA and protein, stress granules are subcellular aggregates associated with neurodegenerative diseases, such as amyotrophic lateral sclerosis (ALS) and frontotemporal lobar dementia (also known as frontotemporal dementia) (Figure 2a, 2b).
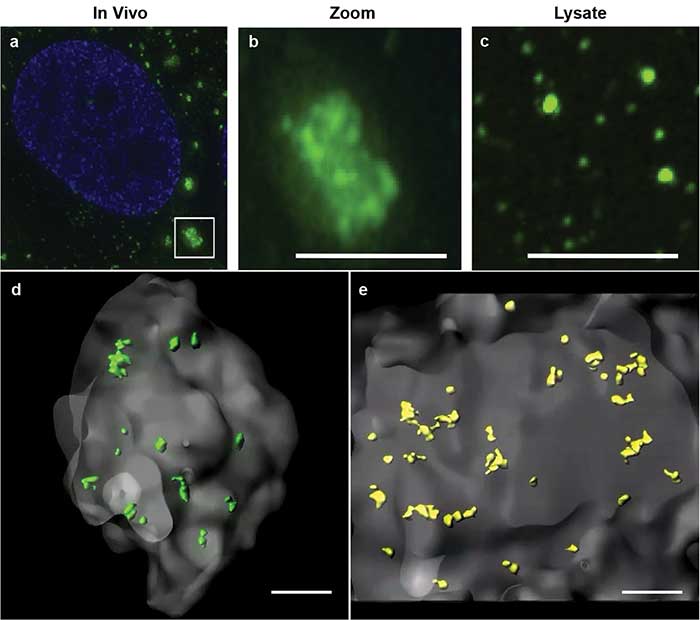
Figure 2. Stress granules imaged by superresolution microscopy. Stress granules (green) imaged by structured illumination microscopy in a cell (a). Zoomed inset of in vivo stress granule (b). Biochemical isolate showing smaller substructures (c). 3D models of double helix superresolution imaging showing stress granules (gray) with dense core structures of protein, GFP-G3BP (green, d) and poly(A+) RNA (yellow, e). All scale bars = 0.5 μm. Images reprinted with permission from reference 4.
Based on biochemical studies, the researchers hypothesized that the stress granules may contain dense core structures (Figure 2c). However, they were unable to easily visualize or measure the stress granule cores in conventional 2D superresolution imaging. Using double helix light engineering, they were able to visualize, identify, and quantify these core structures in three dimensions (Figure 2d, 2e).
Core structures
These quantifications demonstrated for the first time that stress granules contain smaller dense core structures in vivo. Identifying the core structures helped researchers modify their prevailing hypothesis on how stress granules form, dissemble, and relate to pathological aggregates.
These principles of improved depth and resolution can aid researchers in a range of biomedical and materials applications. In the case of single-particle tracking, using the DH-PSF method improves the depth over which a particle can be tracked, enables 3D measurements, and improves the duration over which a particle can be observed.
Traditional methods of particle tracking, such as total internal reflection fluorescence (TIRF) microscopy have very limited axial depth. Consequently, particles diffusing vertically are rapidly lost to defocus and can therefore be tracked only for a limited amount of time. Additionally, particles in biological and biochemical systems moving in three dimensions means diffusion coefficients can be estimated from the 2D data but not accurately measured. With the addition of high-precision axial information and extended depth of field, researchers can obtain more data with more accurate information to directly measure 3D movement.
Wang, Agrawal, and colleagues used DH-PSF technology to improve the signal-to-noise ratio in measurements of chemical interface interactions5. Using the DH-PSF, the researchers were able to track the movement of biomolecules on liquid interfaces6. It was hypothesized based on 2D tracking data that biomolecules take hops in the direction perpendicular to the interface before binding. However, these 3D hops had not been visualized or quantified.
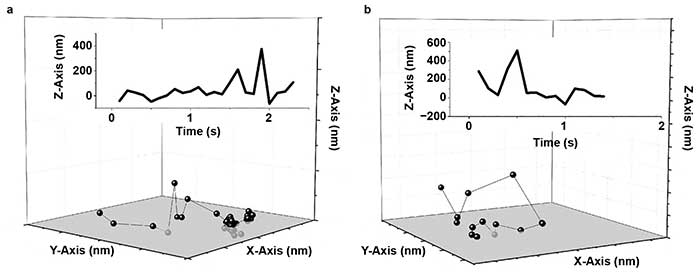
Figure 3. Human serum albumin (HSA) hops on a fused silica (FS) surface. Representative trajectories for HSA on a modified FS surface; X-, Y-, and Z-axis are 2000 nm (a, b). The insets show the corresponding trace of Z position versus time. Reprinted with permission from reference 6.
Wang, Wu, and colleagues demonstrated for the first time that fast 3D molecular hops occurred and, in some cases, flights through the liquid phase were followed by binding at the solid surface (Figure 3)6. In this case, the addition of high-precision axial information was vital to observation of these hopping and flight events.
Developments in light
With high-precision 3D information, the researchers were able to visualize and measure the effects of long-range electrostatic interactions on biomolecule dynamics. This kinetic information demonstrates how proteins move in different biochemical environments and aid in the development of more efficient pharmaceutical purification processes.
Another recent development in light microscopy is the use of planar illumination light, called light sheet or selective plane illumination microscopy (SPIM). Unlike traditional illumination methods, a light sheet illuminates a thin section of the sample that is orthogonal to the detection path. The light sheet reduces background, increases the signal-to-noise ratio, and reduces photobleaching of the sample outside the light sheet.
However, conventional light sheet methods do not allow imaging with high-numerical-aperture (NA) objectives close to the coverslip. These high-NA objectives are required for high-resolution, single-molecule imaging methods such as SMLM. To combine the powerful methods of light sheet and high-precision 3D SMLM, Gustavsson and colleagues created a technique called TILT3D7.
TILT3D uses a tilted light sheet created with a glass prism to image cells close to the coverslip. The setup alleviates the need to have two high-NA objectives in close proximity or submerged in the sample media. It allows for the typical benefits of light sheet imaging, including reduced photobleaching, and is inexpensive to implement.
The increased depth of the DH-PSF captures more microtubules with high-precision axial information over a larger volume than conventional 2D SMLM.
However, this setup results in an angled illumination plane and a thicker (2-µm) light sheet. The extended depth of DH-PSF enables molecule localization over this thicker illumination volume (Figure 4a). This enables precise localization of the molecules within the angled volume and accurate reconstruction of the axial dimension. The addition of the DH-PSF for high-precision 3D localization enabled extremely high-depth, high-precision reconstructions with axial stitching.
Gustavsson and colleagues also used a second modified point spread function with a very large depth range called the tetrapod PSF. A fiducial marker was imaged with the tetrapod to correct for drift and provide an accurate measurement of the axial position of the sample for axial stitching (Figure 4b). With this optical setup, the researchers were able to image and reconstruct structures throughout whole mammalian cells, such as mitochondria and the nuclear lamina (Figure 4c). By combining light sheet, SMLM, DH-PSF, and the tetrapod PSF, Gustavsson and colleagues created a powerful method with improved 3D spatial resolution, a large axial volume, and reduced photobleaching. This method will enable researchers to more easily visualize subcellular structures throughout the entire cell volume with very high precision.
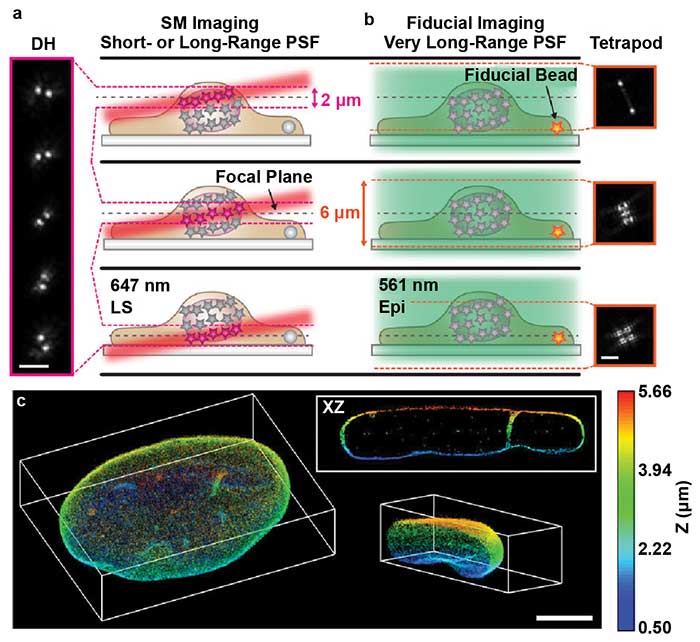
Figure 4. TILT3D combines light sheet microscopy, localization microscopy, and light engineering. Single-molecule (SM) localization with double helix (DH) and light sheet (a). Conventional wide-field imaging set up with tetrapod for drift correction and tracking (b). 3D reconstruction of the nuclear lamina of a HeLa cell, depth encoded in color, scale shown on the right (c). PSF: point spread function; LS: light sheet; Epi: Epi-illumination. Scale bars = 3 µm in (a) and (b); 5 µm in (c). Images reprinted with permission from reference 7.
DH-PSF can be implemented on a variety of microscopes, cameras, and experimental setups. Researchers can accomplish this by using a simple add-on module, such as the SPINDLE, which was designed to integrate with commercially available scientific microscopes and cameras. It works by inserting a phase mask chosen from the library of masks optimized for various axial ranges, emission spectra, and applications. For 3D localization microscopy and 3D particle tracking, the SPINDLE can be combined with localization software such as an ImageJ plugin 3DTRAX. This flexibility allows use of existing setups to perform high-precision, high-depth, 3D measurements without expensive modifications.
Meet the authors
Katie Heiser, Ph.D., is a senior research scientist and applications specialist at Double Helix Optics LLC. She has a doctorate in molecular and cell biology with expertise in high-resolution fluorescence microscopy; email: [email protected].
Leslie Kimerling is co-founder and CEO of Double Helix Optics LLC, a 3D imaging company based in Boulder, Colo. A serial entrepreneur, she has led multiple technology startups from launch through growth. She holds a master’s degree in economics from Stanford University and an MBA from the Anderson School of Business at UCLA; email: [email protected].
Acknowledgments
Some of this material is based upon work supported by the National Science Foundation under Grant #IIP-1059286 to the American Society for Engineering Education. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the National Science Foundation or the American Society for Engineering Education.
The imaging work was performed at the BioFrontiers Institute Advanced Light Microscopy Core. Microscopy was performed on a Nikon Ti-E microscope supported by the Howard Hughes Medical Institute.
References
1. M. J. Rust et al. (2006). Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nat Methods, Vol. 3, Issue 10, pp. 793–796.
2. G. Shtengel et al. (2009). Interferometric fluorescent super-resolution microscopy resolves 3D cellular ultrastructure. Proc Natl Acad Sci, Vol. 106, Issue 9, pp. 3125–3130.
3. S. R. P. Pavani and R. Piestun (2008). High-efficiency rotating point spread functions. Opt Express, Vol. 16, Issue 5, pp. 3484–3489.
4. S. Jain et al. (2016). ATPase-modulated stress granules contain a diverse proteome and substructure. Cell, Vol. 164, Issue 3, pp. 487–498.
5. D. Wang, A. Agrawal, et al. (2017). Enhanced information content for three-dimensional localization and tracking using the double-helix point spread function with variable-angle illumination epifluorescence microscopy. Appl Phys Lett, Vol. 110, p. 211107.
6. D. Wang, H. Wu, et al. (2017). Three-dimensional tracking of interfacial hopping diffusion. Phys Rev Lett, Vol. 119, p. 268001.
7. A. K. Gustavsson et al. (2018). 3D single-molecule super-resolution microscopy with a tilted light sheet. Nat Commun, Vol. 9, Issue 1, p. 123.