A new computational imaging framework enables unprecedented speeds for tissue analysis by mixing cost-effective microspectroscopy with modern algorithms.
HILTON B. DE AGUIAR, ÉCOLE NORMALE SUPÉRIEURE, PSL RESEARCH UNIVERSITY
Conventional bright-field microscopy is the standard for biomedical imaging because it provides cost-effective clinical analysis. However, for researchers and clinicians to assess tissue images accurately, years or even decades of experience are needed. One way to increase selectivity and specificity in life science microscopy is to exploit exogeneous labels; for example, by using fluorophore dyes to enhance contrast in histopathology. Exogenous labels have various drawbacks, as they may perturb the system — especially when used for in vivo applications — and they are not universal, in the sense that laborious tests and developments have to be performed before validating a label methodology.
The Raman effect provides high chemical selectivity with superb optical resolution in label-free imaging. It uses the inherent fingerprint of molecules’ vibrational spectrum. The vibrational spectrum can be thought of as a “barcode” of molecules, with each species having a unique set of peak amplitudes and frequencies in the vibrational spectrum. Therefore, by reading out a vibrational spectrum, one can identify chemical species. A key advantage of Raman spectroscopy is that it uses visible light, therefore providing high-resolution microscopy capabilities on the order of hundreds of nanometers. Classical vibrational spectroscopy, however, uses infrared radiation absorption that, because of its longer wavelength, does not provide high-resolution microscopy capabilities, with resolution to only a few micrometers.
It has been widely demonstrated that Raman spectroscopy provides powerful information for biomedical applications1, including real-time tumor surgery imaging, breast cancer diagnostics, detection of Alzheimer’s plaques, and histopathology. These applications detect and quantify various biomolecules through vibrational spectroscopy analysis, such as proteins, lipids, and nucleic acids. All of these examples present possibilities for using Raman-based imaging, with the majority of proof-of-principle experiments being performed ex vivo or with the sample spatially frozen. However,
in biology, nothing is really fixed in space; because of inherent organelle movement, or just a simple heartbeat, bioimaging often needs to be faster than any movement artifacts. So speeding up Raman imaging is a major challenge tackled by numerous groups worldwide.
Speed vs. expense
In general, there are two types of Raman imaging: spontaneous and coherent (Figure 1). The spontaneous Raman effect typically leads to long acquisition times for a single image, from hours to days. If organelles in a cell are not somehow fixed, images can show smearing artifacts. However, the cost of a spontaneous Raman apparatus could be considered an advantage; it only needs a conventional spectrometer with cheap laser sources. Despite the long acquisition times needed, the simplicity in the spontaneous Raman effect makes it widely popular in applications where speed is not an issue — for example, in airport security checks for remotely detecting chemicals. Conversely, coherent Raman scattering microscopies — with the three most popular versions being stimulated Raman gain (SRG) or loss (SRL) and coherent anti-Stokes Raman scattering (CARS) — have shown much quicker imaging speeds and are able to image at speeds surpassing the ultimate capability of the human eye. However, the price is the infrastructure required; coherent Raman imaging demands costly ultrashort laser sources. Another issue is that coherent Raman imaging typically focuses on small spectral regions; therefore, it does not fully incorporate the barcode complexity of the vibrational spectrum. Recent efforts have advanced the spectral window coverage of this technique2.
Regardless of the type used, to be effective in real-life applications, spectral imaging must overcome a major challenge: data throughput. For instance,
4 GB/s would be the data bandwidth needed for spectral imaging of 512 spectral bands at VGA (video graphics array) resolution. Given the pace of advancements in computer bus bandwidth, it would be a challenge to process useful information in real time. Compressive Raman imaging is an advancement in Raman spectroscopy that allows researchers to speed up the measurement process and concomitantly simplify the data analysis.
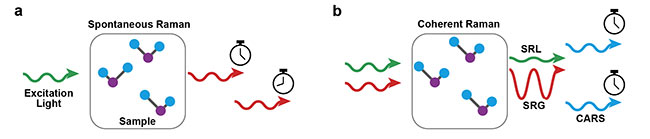
Figure 1. Two types of Raman-based processes. In spontaneous Raman scattering (a), a laser beam impinged on the sample undergoes an inelastic scattering process to generate a new color. The difference between the two colors (in and out) provides the chemical information: the vibrational spectrum. In coherent Raman scattering (b), the molecular vibrations are driven by laser beams with well-defined timings (in opposition to the spontaneous case), which yield extremely strong signal levels and therefore faster imaging speeds. Courtesy of Hilton B. de Aguiar.
Compressive Raman: fast, cheap
To understand why the compressive Raman imaging framework allows for simpler and faster workflow, two distinct recent concepts need to be explained: compressive sensing and single-pixel imaging. A single-pixel camera is much cheaper than a conventional multipixel camera because the price scales with the number of pixels.
Compressive sensing uses efficient computational algorithms to speed up data acquisition. Classically, the frequency of signal sampling for lossless data acquisition is given by the Nyquist-Shannon sampling theorem. That is, to resolve a spectral feature with a dω bandwidth within a spectral interval Dω, one needs Nw = Dω/dω spectral samples. In addition, spectral imaging requires Nw × Ns total samples (Ns = the number of spatial pixels). Compressive sensing works well because many real-life data sets are sparse; some spectral or spatial features are essentially zero, or redundant, and larger imaging would not provide more spectroscopic detail. If this is the case, one can design an experiment with fewer samples than Nw (spectroscopy only) or Nw × Ns (spectral imaging) and then use efficient computational algorithms to fill in the data that was not taken.

Figure 2. A comparison between conventional and compressive spectrometers. In compressive
spectrometers, the costly CCD camera is replaced by a DMD combined with a single-pixel detector.
This scheme is considerably cheaper than the conventional one and may allow for faster imaging. Adapted with permission from Reference 8.
Compressive sensing is largely based on a process called “multiplexed measurement”3. A signal is mixed, detected by a multichannel or even a single-channel sensor, and unmixed computationally. This framework is called “single-pixel imaging” because the original proof-of-concept experiment was aimed at imaging. In contrast, some applications in compressive Raman imaging are based on the principle of single-pixel spectroscopy (Figure 2). A single-pixel spectrometer is equipped with a fast spectral modulator based on a digital micromirror device (DMD). DMDs can operate at high speed (typically tens of kHz), and therefore are compatible with bioimaging applications.
Supervised vs. unsupervised
Reconstructing data that was not originally measured demands prior knowledge of the sample being evaluated. Depending on the level of knowledge available, one can classify the methodology as supervised or unsupervised imaging, analogous to machine learning techniques. “Supervised imaging” means there is strong prior knowledge about the system. In the context of spectroscopic imaging, it means the spectra of pure chemicals are known, and one only wants to know whether a chemical is present or how much of it is present in an unknown mixture. Conversely, “unsupervised imaging” means less prior information is known, and there may be a sparse representation of the data.
Compressive Raman imaging has its origins in a seminal work by a Purdue University group for supervised imaging. In 2013, Wilcox and colleagues4 demonstrated that an optimally designed spectral filter, used with the DMD, can lead to estimation of chemical proportions in a mixture. The basic idea is that, if one measures the pure spectra of the complex mixture beforehand, one can estimate the relative proportions of each species of an unknown mixture (of the same chemicals) using simple optimization routines. Historically, such an approach was inspired by the so-called multivariate analysis, but it still required sampling a complete spectrum. However, Wilcox and colleagues showed that this type of analysis could be implemented directly during the measurement, instead of using the well-known procedure of post-processing with computers. This was a remarkable achievement, culminating in extremely fast Raman imaging speeds by using just a “pinch of photons.” A recent review5 provides comprehensive details about the supervised imaging framework. Nevertheless, supervised imaging has serious drawbacks for biological tissues because the spectral library needed can be large, given the chemical complexity of biological systems.
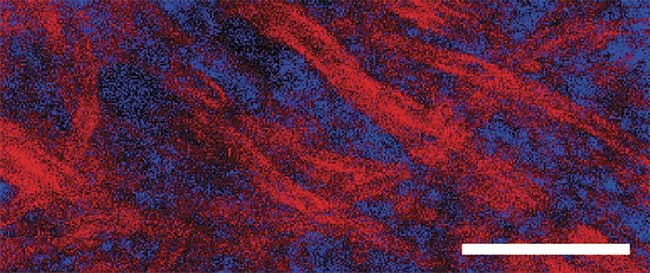
Figure 3. An example of compressive Raman bioimaging. Red represents lipid-rich regions, and
blue represents protein-rich regions. The sample is a brain tissue with observable myelin stripes
surrounding protein-rich regions. This image was taken in a few tens of seconds. Scale bar: 20 µm.
Adapted with permission from Reference 8.
Unsupervised imaging is much less documented, given the more challenging mathematical problem it presents. While supervised imaging is used for classification and quantitation of few variables, in unsupervised imaging, one retrieves the library spectra and their relative amount. This is a computational challenge, and it becomes increasingly more complex as more data points are skipped. Yet, some groups have addressed this problem using classical compressive sensing algorithms. The basic assumption here is that the data has a lot of zeroed points in the spectrum and/or space. That is, one finds data sparsity. The computational reconstruction then seeks a sparse solution through mathematical constraints. This basic sparsity assumption has been used throughout many different fields, including wide-field visible and terahertz imaging3, and more recently in stimulated Raman scattering6.
While unsupervised imaging may be very appealing, classical compressive sensing algorithms are slow. This, in effect, leads to prohibitive reconstruction times if used in high-throughput applications, such as large-scale high-resolution imaging, or dynamic imaging where multiple frames are needed to be computationally reconstructed.
Enabling fast bioimaging
A recent alternative approach in unsupervised imaging used the same mathematical formalism of the so-called Netflix challenge. The original problem went as follows: A set of users rated a set of movies. Some users rated some movies, but not all of the movies, therefore leaving some information blank. The reason this challenge can be solved is because there is a pattern in the users’ ratings, meaning there are correlations embedded in the matrix. Mathematically, we can explain all matrix elements by simply combining information of two other, much smaller matrices. This latter aspect is mathematically known as a low-rank matrix.
A similar structure can be applied in spectral imaging. In a 2D representation, it has been known for quite some time that the hyperspectrum matrix is low rank. That means the matrix contains only a few distinguishable spectral signatures. Each spatial point is simply a linear combination of a few spectral signatures. Therefore, one can skip many bits of the hyperspectrum matrix and still be able to predict the unmeasured information, in effect speeding up the imaging process.
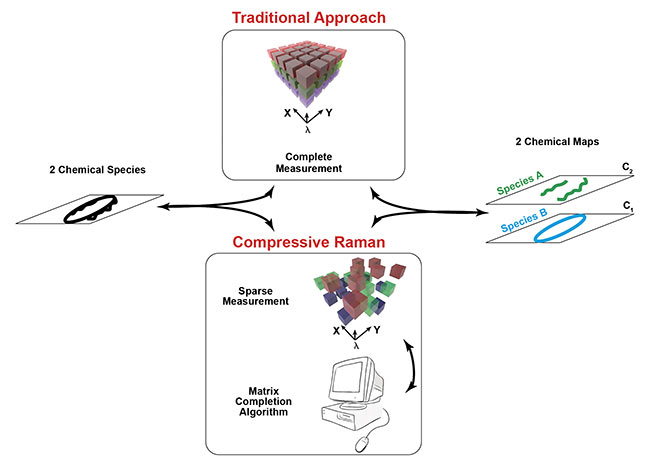
Figure 4. The workflow in traditional and compressive Raman imaging. The traditional approach requires complete measurement of the hyperspectrum
for data representation reduction. In the compressive approach, this data reduction is assumed directly at the measurement level, making the measurement
and the generation of the chemical maps altogether faster.
Using a high-throughput single-pixel spectrometer7, an experiment showed the fastest speeds recorded to date in spontaneous Raman bioimaging (Figure 3)8. The trick was to combine the best of both approaches in compressive Raman imaging by using the reduced data sets of unsupervised imaging, combined with the fast reconstruction speed of supervised imaging. By using the matrix completion algorithm to learn the spectral signatures, researchers could feed supervised algorithms and skip much more data, thus speeding up acquisition and also analysis (see below). By learning the spectral signatures of one tissue sample, the same signature could be used for another tissue, leading to the very same morphological features.
A simpler, faster framework
The compressive Raman imaging framework may enable much higher throughput analysis and could be envisaged to be used in real-time situations.
Compressive Raman imaging not only speeds up the measurement, but accelerates the whole workflow in Raman analysis. Currently, in traditional Raman imaging, a complete hyperspectrum is taken and then post-processed to obtain the final spectral images using tools of chemometrics (Figure 4). This workflow is based on a slow acquisition speed, followed by a post-processing step. In the compressive Raman imaging framework, this chemometrics analysis is done at the measurement stage; therefore, the workflow can be considerably faster. Currently, the only lag time needed is for designing the spectral masks for the programmable spectrometer, but lag time is actually negligible, considering the whole imaging acquisition time. Therefore, the compressive Raman imaging framework may enable much higher throughput analysis and could be envisaged to be used in real-time situations such as chemical sorting (industrial treadmills, such as those used in recycling and pharmaceutical industries), cell cytometry, and histopathology.
Research opportunities
The implications of these recent demonstrations are manifold and could lead to less expensive and more accessible Raman microscopes. In principle, the programmable spectrometer costs less than just one high-sensitivity camera used in traditional Raman setups, which makes the spectrometer much more affordable in developing countries. Reduced electricity consumption could be an additional benefit of single-pixel microspectroscopy.
Compressive Raman imaging allows for cross-fertilization between various disciplines, such as optics, computing science, and analytical chemistry. For instance, in the framework of the matrix completion, we used off-the-shelf algorithms to learn the eigenspectra. Therefore, the development of a dedicated algorithm in matrix completion for the highly correlated features of the hyperspectrum could allow for even better reconstructions, which translates into faster imaging (because the low signal-to-noise ratio in high speed affects the reconstruction), therefore enabling exploration into the faster dynamics of cell biology specimens. On the optical engineering side, most of the setups used in the compressive Raman imaging framework still use relatively simple spatial scanning methods, which could be enhanced with very recently demonstrated efforts, such as using novel modulation schemes9 or combining coherent Raman with compressive Raman concepts10. These examples illustrate how spontaneous Raman imaging may catch up with fast dynamic processes such as chemical reactions and cells and their internal organelles — taking full advantage of the compressive Raman imaging framework.
Meet the author
Hilton B. de Aguiar, Ph.D., is a junior research chair in the Department of Physics at the École Normale Supérieure in Paris. He obtained his doctorate from École Polytechnique Fédérale de Lausanne, Switzerland, where he focused his research on using interface-specific nonlinear spectroscopy tools to unravel molecular-level details of colloidal systems, followed by post-doctoral activities on nonlinear microscopy.
References
1. J.-X. Cheng and X.S. Xie (2015). Vibrational spectroscopic imaging of living systems: an emerging platform for biology and medicine. Science, Vol. 350, Issue 6264.
2. C.H. Camp Jr. and T.M. Cicerone (2015). Chemically sensitive bioimaging with coherent Raman scattering. Nat Photonics, Vol. 9, pp. 295-305.
3. M.P. Edgar et al. (2018). Principles and prospects for single-pixel imaging. Nat Photonics, Vol. 13, Issue 1.
4. D.S. Wilcox et al. (2013). Digital compressive chemical quantitation and hyperspectral imaging. Analyst, Vol. 138, Issue 17,
pp. 4982-4990.
5. D. Cebeci et al. (2018). Recent trends in compressive Raman spectroscopy using DMD-based binary detection. J Imaging, Vol. 5, Issue 1, p. 1.
6. P. Berto et al. (2017). Programmable single-pixel-based broadband stimulated Raman scattering. Opt Lett, Vol. 42, Issue 9,
pp. 1696-1699.
7. B. Sturm et al. (2019). High-sensitivity high-speed compressive spectrometer for Raman imaging. ACS Photonics, Vol. 6, Issue 6,
pp. 1409-1415.
8. F. Soldevila et al. (2019). Fast compressive Raman bio-imaging via matrix completion. Optica, Vol. 6, Issue 3, p. 341.
9. C. Scotté et. al. (2019). Compressive Raman imaging with spatial frequency modulated illumination. Opt Lett, Vol. 44, Issue 8,
p. 1936.
10. H. Lin et al. (2018). Spectroscopic stimulated Raman scattering imaging of highly dynamic specimens through matrix completion. Light Sci Appl, Vol. 7, Issue 5, p. 17179.