Advanced AI algorithms assist researchers in segmenting images and automating image analysis to aid in diagnosis.
GULPREET KAUR AND CHRISTOPHER HIGGINS, EVIDENT Scientific
Improvements in the automation of functional mechanical, electrical, and optical components — along with increasing computing power for image analysis — have enabled the development of high-throughput microscopes, such as the whole slide scanners used in pathology and the fully motorized microscopes used in live imaging and drug discovery. High-throughput systems allow researchers and technicians to use automation to image large numbers of biological samples, resulting in large amounts of data that require trained personnel to analyze. Thus, the backlog in automating the digital imaging workflow has shifted from imaging samples to analyzing images. Artificial intelligence (AI) is now helping to assist in such image analysis — an advancement that has profound implications for the life sciences and medical imaging.
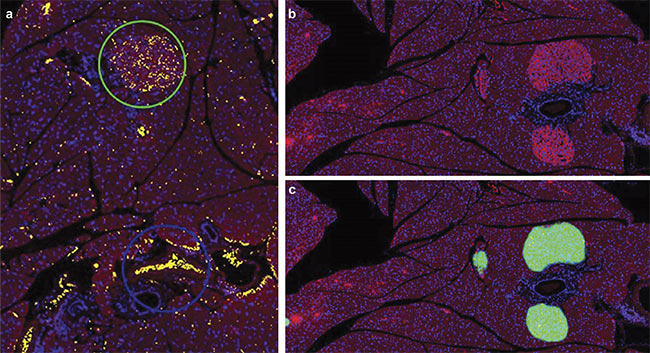
A mouse pancreas section stained with Alexa 594 and DAPI and imaged at 10× magnification using an Olympus SLIDEVIEW VS200 research slide scanner. Slides were stained for insulin-producing beta cells (red) and DAPI (blue). Although a thresholding-based method was unable to differentiate between
erythrocytes (blue circle) and the islets of Langerhans (green circle) (a), AI was able to accurately make this differentiation (b, c). The green mask represents an
AI probability map. Tissue sections courtesy of the Institute for Medical Biochemistry and Molecular Biology/Rostock University Medical Center.
Over the past decade, several free and commercially available algorithms have been developed to enable researchers and clinicians to efficiently segment biological images and identify features of interest. Most such algorithms are based on thresholding, where a cut-off (or threshold) value is defined by the user, and features are selected whose values are measured above or below the threshold. Thresholding algorithms include a combination of properties in their formulae, such as particle intensity, size filtering, and circularity factor. These parameters are useful for segmenting disk-like particles such as nuclei, endosomes, and P-bodies that can help identify specific conditions. A different set of corresponding algorithms is required to recognize and segment spindle-like structures such as cytoskeletons, or branch-like structures such as neurites.
Thresholding-based algorithms require images with a strong signal-to-noise ratio, and these images may require preprocessing through computational methods such as background correction or deconvolution. For the algorithms to function efficiently, prior knowledge of features of interest is required, along with expertise in image analysis to separate the features of interest from the background. In addition, it can be tedious to find an appropriate cut-off value for thresholding algorithms because researchers may differ on what this value should be. Thus, data analysis that uses thresholding algorithms can be subjective, with high user bias in certain situations and low reproducibility. This subjectivity also makes it difficult to automate these data analysis processes for broader use.
Advancements in AI-based algorithms have provided a new and more effective method with which to analyze images. AI-based algorithms designed to analyze biological images have improved image analysis workflows by providing precise segmentation of features of interest and the ability to effectively automate image analysis.
Artificial intelligence
AI involves using computer algorithms to perform tasks that typically require human intelligence to complete. In microscopy and imaging technologies, AI can assist experienced professionals in segmenting images and automating image analysis workflows.
Several AI systems are powered by feature-engineering-based machine learning. Feature engineering applies statistical techniques to recognize patterns — in a series of images, for example — to solve a specific problem without human instruction or complex coding.
Deep learning is a type of machine learning inspired by the structure of the human brain. In AI-based systems, learning occurs as data inputs are processed through interconnected neural network layers, parsed into specialized layers, and mapped to an output layer representing identified features. A convolutional neural network (CNN) is an example of a commonly used deep learning technique that processes data by passing it through an input layer, a set of hidden layers, and an output layer. By processing information within layers and passing progressively refined information between these layers, CNNs accomplish complicated operations with relatively simple processing. Training CNNs requires large amounts of labeled data containing important details, which can require preplanned rigorous procedures and quality controls, as well as dedicated hardware that contains GPUs to reduce processing time. Once trained, though, deep learning systems provide reliable results.
Researchers recently showcased the utility of deep learning methods by successfully identifying and counting glomeruli in kidney tissue slides (Figure 1). Glomeruli are renal structures, or clusters of tiny blood vessels, that are affected by several diseases and conditions. Comparing the numbers of glomeruli in healthy kidneys versus diseased kidneys is of interest in several studies. While glomeruli can be detected by the human eye in a stained kidney section, hand counting each glomerulus is tedious and error-prone. Even thresholding methods have difficulty resolving glomeruli from similar structures. But researchers developed a deep-learning-based segmentation method that was effective at identifying and counting glomeruli.
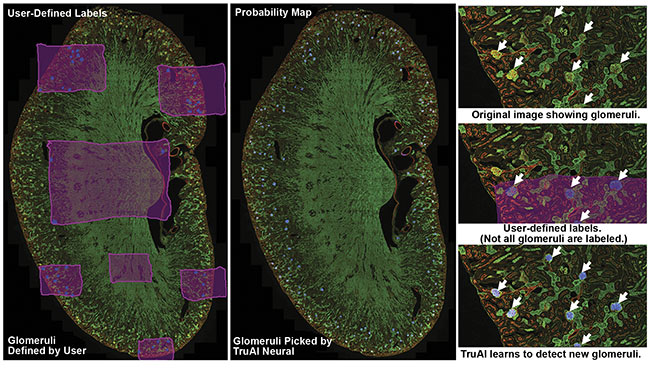
Figure 1. A kidney slide of glomeruli imaged using an Olympus FV3000 confocal microscope. Olympus cellSens software was used to label the background (pink) and glomeruli (blue). The Olympus TruAI deep learning algorithm detected the glomeruli in the kidney section. Courtesy of Olympus Corp.
The workflow used in the example shown in Figure 1 involves three steps. First, the researchers established ground truth by using image analysis software to label images with regions of interest. In this case, glomeruli and the background were hand-labeled. Labeling the background enables the network to better distinguish between different patterns of tissue.
Second, the researchers trained a neural network using the labeled data sets. The AI software develops a probability map that indicates the likelihood of regions of images consisting of glomeruli. During training, AI also uses some of the original data set for validating the developed CNN model. To do this, AI compares training labels with the developed probability map and generates a similarity score to quantify the robustness of the neural network. Similarity scores of 0.8 or higher represent acceptable models (Figure 2).
Third, the researchers developed a neural network to count the identified glomeruli.
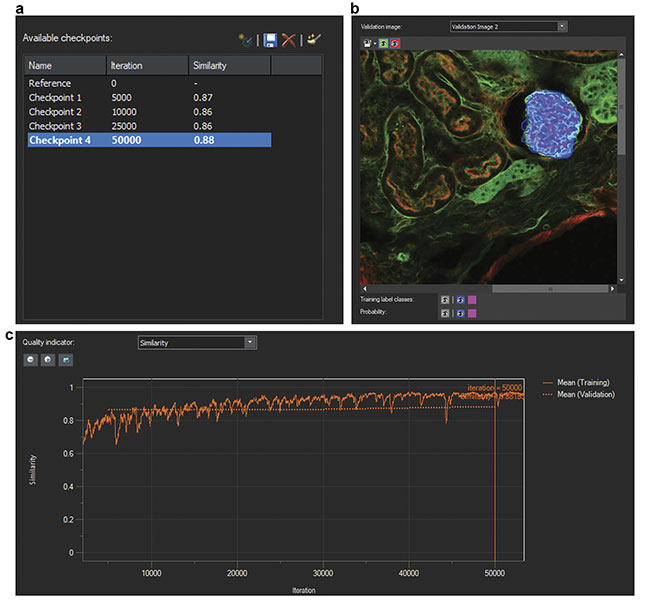
Figure 2. Screenshots of the training of a neural network. The number of iterations and similarity scores (a). A validation image (b). A similarity score indicator over multiple iterations (c). Courtesy of Olympus Corp.
AI in digital pathology
Digital pathology is the process of transforming histopathology slides into digital images, followed by analysis of the digitized images1. To increase throughput, whole slide scanners image slides in a semi- or fully automated fashion. A variety of samples are scanned, including tissue sections, blood smears, and cell suspensions. Samples may be stained with dyes to be imaged using color or fluorescent cameras, or even imaged using polarization to detect birefringent proteins or particles in cells. The image analysis for these samples is similarly wide-ranging and is well supported
by AI.
One AI-based digital pathology technique involves detecting pancreatic islets in a mouse kidney slide. Pancreatic islets are clusters of cells in pancreatic tissue that secrete various hormones. They are easy to detect by the human eye in stained slides. Studying the numbers
of pancreatic islets during the aging process can provide information on diabetes progression2. Mouse pancreas slides were stained for insulin-producing beta cells and DAPI and imaged using a slide scanner (see image at beginning of article). Although a thresholding-based method was unable to differentiate between erythrocytes and the islets of Langerhans, AI was able to accurately make this differentiation.
AI in life science research
Many life science research applications benefit from the use of deep learning algorithms. Examples include detecting punctate structures (nuclei), spindle-like structures (cytoskeletons), and tree-like structures (neurons). When combined with counting algorithms, deep learning algorithms can count particles and events, and they can track the activity of particles over time.
Deep learning has been used in embryonic stem cell research to detect cells and track them over time as they differentiate. Embryonic stem cells undergo rapid cell cycles to proliferate quickly and can undergo cellular differentiation, resulting in the generation of all embryonic layers. Researchers have used a high-content screening system to image embryonic stem cells as they underwent proliferation and differentiation. By combining the screening system with deep learning software, the researchers could identify and track thousands of single cells undergoing changes in shape and morphology. By enabling the high-throughput analyses of images reflecting cell-cycle dynamics, deep learning algorithms enabled a mechanistic understanding of early cell development and reprogramming.
Detecting nuclei is another application of deep learning algorithms because the nuclei can provide an estimated cell count of a feature of interest, enabling the measurement of relationships between varied parameters and numbers of cells. AI-based nuclear detection enables the detection of nuclei with very low light exposure, reducing phototoxicity. Researchers have acquired DAPI images using a high-content screening system set to 100% light intensity, 2% light exposure, 0.2% light exposure, and 0.05% light exposure. They then labeled nuclei in images captured at these various light intensities and used the images to train neural networks. The trained networks proved highly effective at recognizing nuclei in images with low light exposure (Figure 3).
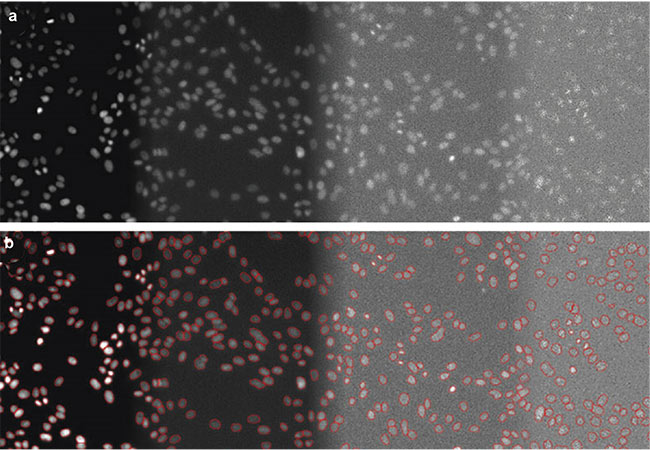
Figure 3. DAPI-stained nuclei of HeLa cells with (from left to right) optimal illumination (100%), low light exposure (2%), very low light exposure (0.2%), and extremely low light exposure (0.05%). The original data (a) and the nuclear boundaries segmented by a deep learning algorithm (b). Courtesy of Olympus Corp.
Deep learning software has also been shown to be effective in detecting unstained nuclei in bright-field images. In addition to preventing the unnecessary exposure of samples to high-energy light and preventing phototoxicity, this method of detection also allows the freeing up of a filter channel (often DAPI) for imaging. This additional channel can be used to perform complicated applications, such as the translocation assay. In this assay, several transcription factors (proteins involved in converting DNA into mRNA), such as the androgen receptor (AR), translocate to active sites at which gene transcription is occurring within the nucleus, where the transcription factors are detectable using a green fluorescent protein (GFP)-AR antibody.
Researchers used a confocal microscope to predict the dynamics of the androgen receptor. The translocation of the androgen receptor was visualized as a distinct speckling pattern. The researchers analyzed the resultant images using image analysis software with a deep learning module to generate a neural network to detect unstained nuclei and differentiate between nuclei with and without speckling. The translocation assay is more complicated than other segmentation assays because it requires the software to identify the location of transcription factors and not just their presence. Deep learning algorithms can easily achieve this.
The ability to image live cells and identify and segment various phenotypes is particularly useful for drug discovery research. Samples housed in 96- or 384-well plates are treated with different drugs, and various phenotypes are investigated. Entire 96-well plates of cells or organ spheroids can be imaged in multiple channels, and different parameters, such as the size/volume of organelles and organelle counts, can be automatically compared to nuclei counts, enabling high-content imaging and analysis.
AI in medical imaging
Several medical fields — including radiology, pathology, ophthalmology, and dermatology — routinely rely on the analysis of imaging data. AI-based approaches facilitate these fields in many ways, including by aiding in diagnosis by detecting and characterizing features on images, and by monitoring diseases over time. AI is good at recognizing complex patterns in images and automatically providing quantitative assessments. AI can be applied to computed tomography (CT), magnetic resonance imaging (MRI), or digital pathology images of biopsy tissues and taught to identify lesions or tumors and to classify them as malignant or benign and primary or metastatic3.
After an internal validation, AI software for medical diagnosis adopts either a first-read or a second-read workflow. In a first-read workflow, AI performs the primary screening and sorts images based on findings, with the physician signing off on the final results. In a second-read workflow, a pathologist reads all of the images in the case and sends it to AI to see whether anything was missed. First-read AI workflows are popular because they significantly speed up diagnosis. In fact, the FDA recently granted De Novo marketing authorization for an AI-based first- or primary-read product (by Paige), and it granted breakthrough device designation (a fast-tracked review) for an AI-based prostate cancer screening module (by Ibex). Pathologists using these technologies train the AI with highly annotated digital pathology slides and use over 100,000 images of each cancer type.
AI can also be used in treatment planning. AI algorithms can learn which treatment plans for radiation treatment, for example, have achieved good outcomes with tumors of a particular size and at a particular location. On a broader scale, AI can provide a holistic view of disease in a patient and can model various outcomes. This ability will improve understanding of a disease and its dependence on various factors.
Hurdles
AI has already gained widespread acceptance in many scientific communities, and this trend is likely to continue.
In life science research and digital pathology, AI can improve the results produced by microscopy and imaging technologies. For instance, slide scanners rely on contrast-based autofocus algorithms to find the area of focus on a slide prior to imaging. Most of these focus-finding mechanisms involve the microscope scanning through various z-planes on a slide and, based on contrast, selecting the plane in focus. This method is prone to failure when the contrast is high, such as when bubbles are in the slide or dust is on the coverslip.
Deep learning systems, on the other hand, can be taught to recognize the focused sample to improve this process of scanning through various planes, making focus-finding quicker and less error-prone.
In medical imaging, AI is gaining acceptance at a cautious pace. The FDA has thus far only cleared or approved AI devices that rely on a “locked” algorithm, and any adjustments to the software would require further review. Two ethical questions that must be resolved are: “Who owns the AI?” and “Who is responsible for the accuracy of the results (the diagnosis and treatment plan)?” By current standards, each diagnosis must be signed off on by a pathologist or physician, even if AI algorithms were used. Such standards are likely to remain in place for now. In fact, many users refer to AI algorithms as “augmented intelligence” since they are primarily being used to assist physicians.
A major concern with using deep learning methods in medical imaging is that they use a series of hidden layers of analysis to generate the neural network during training. No one fully understands how a deep learning algorithm arrives at a given result. Consequently, deep-learning-based methods are often called “black box” algorithms.
Training multiple networks with the same data may lead to different models with the same similarity score. This means that, over time, different interconnections in the hidden layers could lead to different results after the data is repeatedly entered into the same network. This makes it difficult to predict failures or troubleshoot issues. Further research is being done to understand how hidden layers function and to quantify their impact better. Considering this need for better understanding, statistics-based feature-engineering methods have a better chance of being approved by government agencies for automated diagnosis when the results achieved from the methods prove to be consistent.
Once these concerns have been addressed, building comprehensive models to detect abnormalities in the human body should be possible. Many factors are involved in training such networks with information from different demographics involving various conditions. A current major roadblock is the large amount of labeled data required to build such an AI algorithm. This problem is further complicated by the incidences of rare diseases, for which automatic labeling algorithms are nonexistent and subject knowledge is limited to a few specialists who can verify the accuracy of the labels and the results. Building such an “ensemble” model will require machines with very high processing power.
The hope for the future is an AI algorithm trained on an amalgamation of all scientific and medical research, an algorithm that could diagnose conditions as well as suggest action plans for the best probable outcomes. AI could make this task possible, and it would augment the collective intelligence of clinicians and researchers from a broad spectrum of fields.
Meet the authors
Gulpreet Kaur, Ph.D., is a research imaging applications specialist at Evident Scientific, a subsidiary of Olympus Corp. She completed her doctorate in molecular biology at the University of Wisconsin-Madison, where she studied organelle biology using live-cell spinning-disk confocal microscopy. Kaur has more than eight years of experience in microscopy and has worked with various technologies, including confocal, spinning-disk, superresolution, and light-sheet imaging. She served as a trainer for a microscopy and image analysis core facility at the University of Wisconsin-Madison and consulted with scientists on experiment design and optimization of imaging conditions; email: [email protected].
Christopher Higgins is business development manager for clinical and clinical research markets at Evident Scientific, a subsidiary of Olympus Corp. A 25-plus-year veteran of the microscopy field, he has spent much of his career in management of products, sales, and applications for a wide variety of biological light microscopes used in both research and clinical applications. In his current position, Higgins focuses on the understanding, adoption, and expansion of the digital pathology, image analysis, and artificial intelligence fields; email: [email protected].
References
1. M. Niazi et al. (2019). Digital pathology
and artificial intelligence. Lancet Oncol, Vol. 20, No. 5, pp. e253-e261.
2. P. Seiron et al. (2019). Characterization of the endocrine pancreas in type 1 diabetes: islet size is maintained by islet number is markedly reduced. J Pathol Clin Res, Vol. 5, No. 4, pp. 248-255.
3. N. Sharma and L.M. Aggarwal (2010). Automated medical image segmentation techniques. J Med Phys, Vol. 35, No. 1,
pp. 3-14.