Researchers are using photonics to help lower the cost of sequencing the human genome.
Hank Hogan, Contributing Editor
The cost of sequencing an individual’s genome today is at least $10 million, which is much too high to allow studies of the genetic basis for disease or to make genomic sequencing part of standard medical care.
That’s why the National Human Genome Research Institute in Bethesda, Md., recently awarded tens of millions of dollars in grant money for research aimed at cutting the cost of sequencing, first to $100,000 and then to $1000. The expectation is that the intermediate cost will be achieved within five years.
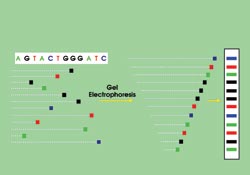
In Sanger sequencing, DNA is randomly snipped from a genome and tagged with fluorophores, one for each type of base pair. These snippets are separated according to length during gel electrophoresis. By reading the fragments, researchers sequence the DNA. Courtesy of Jeffrey A. Schloss, National Human Genome Research Institute.
First developed 30 years ago by double Nobel laureate Frederick Sanger, current sequencing techniques replicate DNA segments by polymerization and incorporate either a radioisotope or a fluorescent label for each of the four bases: adenine (A), thymine (T), cytosine (C) and guanine (G). The solution also includes a low concentration of a modified version of one of four biochemicals that halt the replication when incorporated by the polymerase.

Each spot is a bead bearing immobilized DNA, and its color indicates the identity of the base. By stepping through locations on a DNA fragment, researchers use such fluorescence images to decode the DNA sequence. Courtesy of Gregory J. Porreca and Jay Shendure, Harvard Medical School.
After some preparation, the amplified results, which are of random length, are forced down a gel by a negative voltage that repels the negatively charged DNA. The fragments separate according to length, and, by looking at the tags, researchers can classify the fragments from both length and base type. The result is familiar to anyone who has seen a recent crime drama: four horizontal columns of vertically spaced bars.
The Sanger technique was automated more than 15 years ago, and researchers used these systems to sequence the human genome. The cost to sequence a base pair has fallen by a factor of two every 12 to 18 months or so since the technique was introduced, to the point where the cost is now about a dollar per 1000 bases and the speed about 20 bases per second of instrument time. Experts do not believe that this pace of improvement can be sustained.
“We’re running out of ways to keep achieving those twofold cost reductions because it has been refined over so many cycles,” said Jeffrey A. Schloss, program director for technology development at the institute.
To be sure, the cost reduction will continue for at least a few more cycles and could lead to genome sequencing for $100,000. But even that, experts conclude, might be a stretch. Still, there are a number of promising approaches, many of which are based on optical methods.
At the bottom of a well
A team from 454 Life Sciences Corp. of Branford, Conn., the University of California, Berkeley, Rockefeller University in New York and the Rothberg Institute for Childhood Diseases in Guilford, Conn., reported on a technique that involves an optical array in the Sept. 15 issue of Nature. The technology is roughly 100 times faster than existing sequencing processes.
In this method, sequencing takes place on fiber optic slides. The slides are made by Incom Inc. of Charlton, Mass., by arranging fibers into a bundle, heating it and drawing it out, arranging the bundles into another bundle, and repeating the process. After three or so passes, a hexagonal array of microscopic optical fiber cores surrounded by light-confining cladding make up the final bundle. “You then slice that up, and each of the individual fibers is now perfectly coherently aligned,” said Michael A. Detarando, Incom vice president for product development.
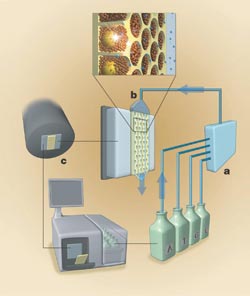
One sequencing instrument consists of a fluidic assembly (a), a flow chamber that includes the well-containing fiber optic slide (b), a CCD camera-based imaging assembly (c) and a computer that provides the user interface and instrument control. Reprinted from Nature with permission of the researchers.
In the next procedure, the core at one end of each fiber is etched away to make a shallow well 44 μm in diameter surrounded by 2 to 3 μm of cladding. Marcel Margulies, vice president for engineering at 454, said that the researchers pressed the unetched end of the bundle against a 50-mm-thick fiber optic faceplate with 6-μm-diameter fibers. The faceplate is permanently attached to a 16-megapixel Fairchild CCD, which allows the imaging of photons emitted from each well.
The researchers filled the wells on the slide with beads bearing single strands of the DNA to be sequenced, carefully selecting the bead size so that it was unlikely that two beads would end up in one well. They then flowed reagents over the slide, looking for and capturing the chemilumenescent flash at about 560 nm that signaled the incorporation of a base reagent by the DNA strand.
Because they know which nucleotide is flowing, they know which base is incorporated, Margulies said. The intensity of the signal determines how many times that base was added.
He noted that an instrument based on this technique is already commercially available, has been put into operation in genome centers and is covered by a recent exclusive distribution license agreement with Roche, based in Basel, Switzerland. Improvements to the device and technique are ongoing. These include shrinking the wells and increasing their utilization above the current 35 to 40 percent. Beyond that, planned enhancements include faster delivery of reagents and reading longer lengths of DNA. Margulies is confident that an improvement by two orders of magnitude is possible as state-of-the-art technology becomes commercially available.
Solving the four-color problem
Another bead-based approach comes from researchers at Harvard Medical School in Boston and Howard Hughes Medical Institute at St. Louis-based Washington University. This technique, the researchers reported in the Sept. 9 issue of Science, has a cost per base of about one-ninth of existing methods and is an order of magnitude faster.
In this scheme, DNA molecules are amplified in parallel on 1-μm-diameter beads. These beads are immobilized and interrogated through the use of fluorescently tagged reagents, one color for each of the four DNA bases. By using the reagents in succession, the researchers get a signal that reveals which base is at a given location. They strip off the tagged reagents and repeat the process for another location. In this way, they sequence the DNA.
Gregory J. Porreca, a graduate student at Harvard Medical School, noted that the technique cannot be used to sequence a genome from an organism that has never been sequenced before. Rather, it is suited to resequencing a genome by looking for differences between a known template and the particular sequence of a given individual. This works because the human genome that was sequenced in 2003 is a template composed from several individuals. It is 99.9 percent identical to anyone’s genetic material.
Although performed with a standard epifluorescence microscope, the technique puts demands on the imaging equipment. Potentially, a billion 1-μm-diameter beads can be spread across a standard microscope slide. “In current Sanger sequencing instruments, the data acquisition rate is the rate-limiting step in the entire sequencing pipeline, and this bottleneck sets the cost for the sequencing method (since instrument time is the primary cost),” Porreca said.
The same limitations will probably exist in the new method. The camera used must operate at 30 frames per second or better and must have a pixel count of at least a million, a detector size of a centimeter or more, and high sensitivity. However, according to Porreca, such state-of-the-art specifications aren’t enough.
“To maximize the data bandwidth the instrument can provide, we would want a larger CCD which covers the objective’s entire field of view of about 22 mm, high gain with low background, high frame rate of 30 fps plus, and pixel size of 8 to 18 μm,” he said.
He added that the expectation is that the technique will cost less than $100,000 per genome. Reaching the $1000 figure will mean acquiring data continuously at greater than 250,000 base pairs per second. That requires operation very near the limit of one CCD pixel per useful DNA feature. That may prove difficult to achieve, but brighter fluorophores and better optics would help.
Zero modes and nanopores
Nanofluidics Inc. of Menlo Park, Calif., is developing a DNA sequencing method based on research originally done at Cornell University of Ithaca, N.Y. The technique uses zero-mode waveguides constructed out of metal films thinner than 100 nm and peppered with holes about 50 nm in diameter. The films are deposited on a coverslip.
Because the holes are smaller than a wavelength of light, the fluorescence that results from the sample comes from a small volume and can be detected despite other fluorescent signals. There’s no need to dilute reactants — an important point because DNA polymerization requires a certain concentration to work. For sequencing, the reactants are put in the bottom of the zero-mode waveguides while fluorescently labeled nucleotides float above in solution. As the fluorescent labels are incorporated, a signal is used to decode the sequence of the target DNA.
David Hanzel, vice president of product marketing at Nanofluidics, won’t reveal details about when the products might be available. But he acknowledges that the goal is the $1000 genome and that there are challenges in the detector, illumination, optical path and real-time data collection. “We don’t have to invent anything, but we’re on the bleeding edge of some of the technologies,” he said.
Another single-molecule, highly parallel approach involves nanopores and single-molecule fluorescence detection. The nanopores act as a sieve, allowing only molecules of a certain size through. A 1.5-nm-diameter hole in a silicon nitride film, for example, would accept a single strand of DNA but not a double helix. Amit Meller, a senior fellow at the Rowland Institute at Harvard in Cambridge, Mass., has come up with a scheme to exploit this.
In Meller’s approach, an analogue of the target DNA sequence, created by the Norwegian company LingVitae AS, is tagged with complementary DNA segments capped by a fluorophore at one end and a quencher at the other. When a negative voltage is applied to one end of a chamber divided by a nanopore membrane, the DNA attempts to wiggle through the holes. The DNA helix unzips and thereby removes the quencher of one segment from the fluorophore of the next. Once through, the molecule folds in on itself and self-quenches. The result is a flash of light, the color of which announces which base has just gone through the nanopore.
In theory, this method promises very fast sequencing at very low cost. Meller noted that it’s still early in the development of the technique, although a proof of principle is under way. He added that there are many photonic challenges, some of which are in hardware and some in software. “We have recently made some great progress toward this goal, but there is a lot of image processing first to determine, for each and every pore, the sequence of flashes,” he said.
Fretting about beads
A final single-molecule approach involves Förster resonance energy transfer (FRET). When a donor fluorophore is within a few nanometers of an acceptor fluorophore, the resulting transfer of energy from donor to acceptor causes emission from the acceptor.
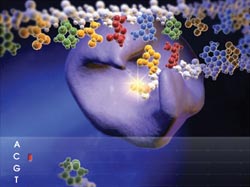
One sequencing strategy detects single-pair FRET between a donor-labeled polymerase and an acceptor-labeled nucleotide during DNA synthesis on a micron-size bead. Running many beads in an array allows for parallel detection. Courtesy of VisiGen.
VisiGen Biotechnologies Inc. of Houston is seeking to exploit this by detecting color-coded acceptors that are attached to the nucleotide as the polymerase builds the complement of the DNA being sequenced. According to VisiGen President and CEO Susan H. Hardin, the method allows for massively parallel detection as arrays of sequencing complexes build completely natural DNA polymers. The company’s plan is to follow the sequential addition of each nucleotide in real time by detecting spectral shifts as acceptor and donor fluorophores are brought into and removed from proximity to each other.
For competitive reasons, Hardin won’t go into many details. She said that VisiGen engineers the modified nucleotides so that the acceptor fluorophores are in the right place. The company also makes the polymerase so that the donor fluorophore is positioned correctly. She said that sequencing surfaces based on this idea are expected to be offered by the end of 2007.
Although many of the challenges to this approach are rooted in organic chemistry, some involve characteristics that many single-molecule researchers crave. “People always want fluorophores to become more stable, brighter and better resolved spectrally. Additionally, you want to have very, very sensitive CCD chips,” Hardin said.