The olfactory system that lets mammals detect and identify odors starts with a wide array of G-protein-coupled receptors (GPCRs), which are located on olfactory sensory neurons (OSNs) that comprise the olfactory epithelium, a thin layer of tissue deep within the nose. This network has the capacity to identify and respond to the host of odors that mammals, including humans, come across every day.
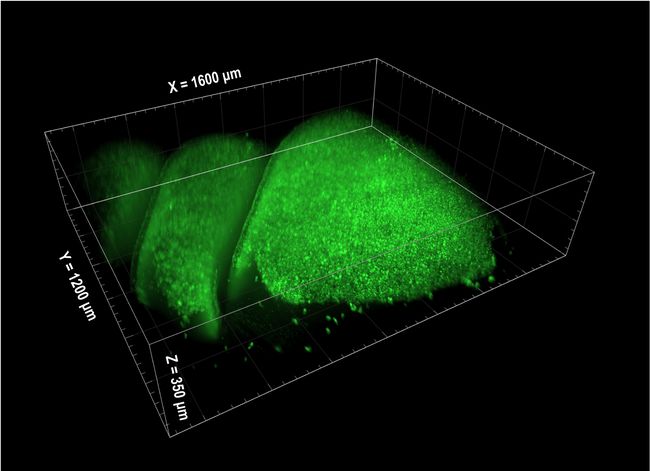
A 3D SCAPE image of an olfactory epithelium in a mouse. Courtesy of Hillman and Firestein Labs.
A group of researchers has unlocked just how this process works, thanks to the use of advanced optical technology.
In its recently published research, a team from Columbia University documented the simultaneous imaging of the responses of thousands of OSNs within the mouse nose by using a high-speed imaging method called swept confocally aligned planar excitation (SCAPE) microscopy.
By using calcium-sensitive fluorescent proteins, the team extracted each cell’s responses to a sequence of individual or mixed odors. The odors included almond, floral/jasmine, and citrus. Whether these odors were introduced individually or at the same time seemed to have a profound impact on the way the neurons responded to them.
The researchers learned that one odor could actually enhance or suppress a neuron’s response to a second odor, even when the neuron did not respond to the first odor when delivered alone. They proposed that these complex interactions within the nose may help the brain to identify a much larger range of odor mixtures compared to trying to separate and identify individual components of a blend of odors.
The study was made possible by the use of SCAPE microscopy, a technique developed in the laboratory of Elizabeth M.C. Hillman, professor of biomedical engineering and radiology and principal investigator at Columbia’s Zuckerman Institute. She said SCAPE is a form of light sheet microscopy that provides very high-speed 3D imaging of large, living, intact samples through a single objective, and also has much less photobleaching than methods such as confocal microscopy.
“SCAPE is very efficient with its use of light, since photons get many chances to excite fluorescence as they travel along the direction of the sheet,” Hillman said. “The use of a camera permits parallel measurement of all of the pixels in the plane at once, greatly increasing integration time per pixel, permitting lower overall illumination levels.” Hillman’s group has built over 20 SCAPE systems to date and licensed the technique to Leica Microsystems for commercial development.
Stuart Firestein, professor of biological sciences at Columbia University and a senior author of the study, said the next step in their research will be to learn how odor combinations are translated into activity in the brain. The team said its work could have implications in monitoring responses to various drugs and unlocking some of the mysteries of Alzheimer’s and Parkinson’s diseases, of which loss of smell is a symptom.
In ongoing projects, the researchers plan to expand characterization of these effects and to study animals with genetically altered receptors.
“We are also sharing SCAPE widely with researchers working in a wide range of different samples — from small organisms like C. elegans worms, zebrafish larvae, fruit flies and their larvae, etc.,” said Hillman. “We are also imaging the awake mouse brain using single- and two-photon versions of SCAPE and are even imaging bubble and crystal slurry dynamics in 3D in collaboration with a volcanologist. We are developing a medical version of SCAPE for in vivo, in situ histopathology, and we have shown that SCAPE can also do very high throughput structural imaging of cleared and expanded tissues.”
The research was supported in part by the Brain Research through Advancing Innovative Technologies (BRAIN) Initiative within the purview of the National Institutes of Health (NIH). Edmund Talley, program director at the National Institute of Neurological Disorders, said one of the main goals of the BRAIN Initiative is to develop technology for investigating the role of neural circuits in health and disease.
Like other projects, the work of the Columbia research team was judged on a number of criteria for funding, he said.
“NIH applications are subject to two levels of review, first by scientific peers, where they are evaluated for their scientific merit, and then by the institute advisory councils,” Talley said. “For the BRAIN Initiative, grants are also informed by the guidance of the Multi-Council Working Group.”
Other support for the project was provided by the National Institute on Deafness and other Communication Disorders, the National Cancer Institute, the U.S. Department of Defense, a Firmenich contract, the Simons Collaboration on the Global Brain, the Kavli Institute for Brain Science, and the National Science Foundation.
The research was published in Science (www.doi.org/10.1126/science.aaz5390).