For clinicians and patients to benefit from rapid Raman-based point-of-care analysis, a system must analyze small amounts of fluid with specificity, using AI tools to include important data in the diagnostic process.
ANJA SILGE AND JÜRGEN POPP, LEIBNIZ INSTITUTE OF PHOTONIC TECHNOLOGY
Raman spectroscopy could soon aid traditional blood diagnostics at the patient’s bedside. Courtesy of iStock.com/Jacob Wackerhausen.
Raman spectroscopy offers new possibilities for blood diagnostics, which is crucial for making timely decisions in the intensive care unit (ICU) (Figure 1). Progressions in clinical knowledge and well-defined objectives of a technology’s use guide developers toward inventive technical opportunities in a clinical setting. This synergy enables the translation of insights from both fields into groundbreaking diagnostic processes. By merging clinical expertise with the latest instrumentation, developers can create more accurate and efficient diagnostic tools. A continuous sample-to-answer approach with a Raman spectroscopic readout is particularly well suited for this task, because it measures a blood sample’s components, and after proper sample preparation, puts the right biological targets in the laser focus.

Decisions must be made quickly at the point of care in the intensive care unit (ICU). Courtesy of iStock.com/SDI Productions.
Infections — many of which manifest in the blood — frequently lead to critical and life-threatening conditions in the ICU. Clinicians may see patients with very similar symptoms that actually have varying illnesses. As a result, two key strands of information must be tracked and incorporated into a diagnosis in a timely and effective manner. The first of these strands includes the cause of the symptoms, which can be determined by identifying whether the infection is viral or bacterial. This distinction is crucial, leading to the use of distinct therapeutic approaches.
For bacterial infections, it is essential to know the most effective antibiotic and its concentration, because this will influence the treatment. It is therefore crucial to have access to advanced tests that are precise, sensitive, and quick to prevent the unnecessary, inappropriate, or ineffective use of antibiotics. With growing trends in antibiotic resistance, it is vital to provide this treatment only when necessary1.
The technological developments in clinical Raman spectroscopy have been oriented toward unmet clinical needs and the challenges in obtaining therapy-relevant information.
The second strand of information includes the distinct clinical phenotypes and host-response patterns of the patient, which require individual therapeutic approaches for each condition. Substantial advancements in basic science, in combination with extensive clinical or epidemiological studies, have increased the medical community’s knowledge of life-threatening organ dysfunction caused by a dysregulated host response to infection. This condition is known as sepsis. The advanced knowledge about sepsis must be translated into new diagnostic processes to accelerate therapeutic interventions and improve patient outcomes. Critical information must be captured to intervene safely and reliably at the right place and time1.
Developers are now tasked with connecting clinical diagnostic gaps using intelligent solutions. The aim is to adapt the advantages of new technologies, such as Raman-based in vitro diagnostics, to the latest medical findings. What does it mean for a developer when a clinician requests information on the clinical phenotype behind symptoms and the appropriate antibiotic in a timely manner? To be fast, the information must be obtained from easily accessible patient material, such as biofluid that may have already been collected (opening image). The processing of patient material should be achievable in a few straightforward steps. And this process needs to be automated.
It is necessary to extract information from small sample volumes to operate on-chip Raman spectroscopy, which entails reading data from miniature devices2. Panel 2 of Figure 2 depicts designs of such a sample container with multiple sampling units in the size of a microscope slide and a miniaturized Raman instrument as developed at the Leibniz Institute of Photonic Technology eV (Leibniz-IPHT). This enables the instruments to be positioned near the patient’s bedside, and, if properly implemented, testing can be completed robotically.
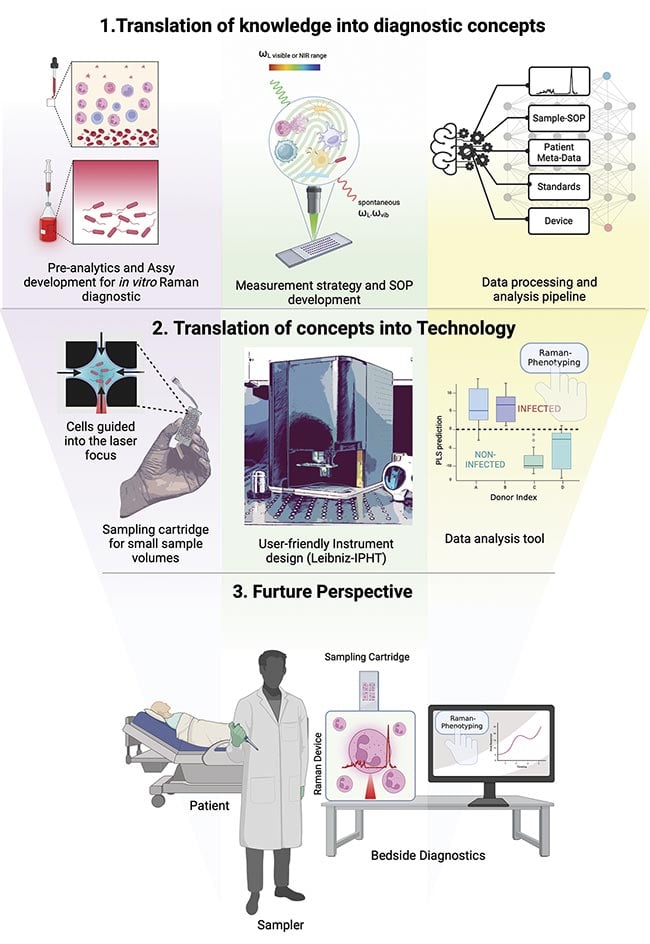
Figure 2. The mission of developers in the field of clinical Raman spectroscopy. Illustration created with BioRender.com; photo of the sampling cartridge and Raman instrument courtesy of Sven Döring/ Leibniz-IPHT.
Blood diagnostics are an important and objective source of information about an infection and the immune response, and they are easily accessible in clinical settings. Blood circulates continuously, and the analysis of its components by Raman spectroscopy offers immediate insights into a person’s health and medical conditions. As a result, blood cells are easily obtained for diagnosis and contain important information about the patient’s immune response. In the ICU, it is of paramount importance to test bacteria that can affect the entire body, which are identifiable in the blood. Blood cultures are essential in this context; the bacteria are grown in a culture medium inoculated with the patient’s blood to test them against various concentrations of antibiotics1.
Raman-based diagnosis
Raman spectroscopic diagnostics offer a straightforward, objective, and label-free approach to sampling small volumes of biological material. Raman microspectroscopy is a technique that allows for the analysis of small samples at a microscopic level. Using lasers with high precision enables the analysis of the molecular composition of any biological structures, such as blood cells at a resolution of a few micrometers, which converts the data into diagnostic information directly. Since a few microliters of blood contain thousands of cells, a representative cell population can be measured. Collecting such a small volume is minimally invasive for patients, because it does not require significant discomfort or inconvenience during extraction.
Figure 2 outlines the overall goals of developers in the field of clinical Raman spectroscopy. The first stage is to translate the knowledge about Raman spectroscopic applications into diagnostic concepts. Practitioners of clinical Raman spectroscopy have established approaches for comprehensively investigating cell samples. The pre-analytics stage, which encompasses comprehensive and expedient sample preparation, reveals all pertinent phenotypic data. All technological necessities for sample presentation for subsequent Raman microscopic analysis are considered.
Researchers from Leibniz-IPHT, the Jena University Hospital in Germany (the Center for Sepsis Control and Care, Department of Anesthesiology and Intensive Care Medicine, Institute for Clinical Chemistry and Laboratory Diagnostics), the internal medicine department of the National and Kapodistrian University of Athens in Greece, and the clinical research center of the Copenhagen University Hospital in Denmark have demonstrated that the spectral patterns of white blood cells can indicate various subtypes and activation states3. This analysis has considerable potential because it facilitates the acquisition of crucial, unbiased information about infections and immune responses through the direct sampling of blood cells without the need for labeling or additional processing.
On this basis, it is essential that physicians conduct multicenter trials to fully understand the value of the technology and the improvement in patients’ outcomes. This will help them to identify the ranges in Raman data derived from normal and affected populations, and to determine the diagnostic sensitivity/specificity across different studies. To achieve this, it is crucial that technology is translated into user-friendly instruments and that comprehensive sample-to-answer workflows are developed.
Data analysis
Experimental research carried out with partners from Leibniz IPHT and Jena University Hospital has demonstrated that machine learning can distinguish between various infections caused by bacteria, viruses, or fungi using the Raman signatures of white blood cells4,5. The sample’s Raman data are computationally associated with all essential metadata, including biological and technological data. This integration ensures that the data is accurately transformed into useful diagnostic information with respect to all technological pitfalls6. The development of diagnostic tests for critically ill patients has demonstrated that the Raman signatures of blood cells can provide crucial details for the identification and classification of various medical conditions, including sepsis, which is essential for the selection of appropriate treatment3,7.
Another crucial aspect of blood diagnostics is the processing of blood cultures. As previously discussed, bacterial bloodstream infections are highly dangerous due to their rapid spread throughout the body. The most critical information for clinicians is the selection of the most effective antibiotic in the appropriate concentration to fight these bacteria. This assertion can be most effectively conveyed through phenotypic resistance tests. This entails testing the isolated bacteria from the blood against specific antibiotic concentrations, thereby initiating a targeted antibiotic therapy.
To conduct these tests, standardized bacterial suspensions with a culture density of 108 colony-forming unit (CFU)/mL are required. This culture density is achieved within a standardized blood culture through a 12-h incubation period. The bacteria must then be dispersed over several test units to grow against various antibiotics for at least 12 more hours. The “yes/no” statement regarding growth is converted into an antibiogram (an overall profile of antimicrobial susceptibility), which assists the clinician in determining the most appropriate treatment1.
Methods that could perform these phenotypic resistance tests on early micropopulations, distributed over a relevant antibiotic test panel, would provide significant advancement. Raman spectroscopic resistance testing can play an important role in accelerating this process. Firstly, the cell populations required for Raman testing can be significantly reduced. For Raman-based antibiotic sensitivity testing, a few hundred cells in the laser focus are sufficient2,8. Researchers are currently trying to distribute this number of cells evenly in each test unit and to get them effectively into the laser spot. The aim is to reduce the number of CFU/mL from 105 to 103. This will result in a significant reduction in the cultivation time. For the resistance test, the bacteria only need to be exposed to the antibiotic for 90 to 120 min. Specific changes in the spectrum captured with Raman instrumentation can reveal whether a bacterium is resistant or sensitive2,8.
This rapid turnaround time allows for the identification of antibiotic resistance patterns in a timelier manner, thereby enabling health care providers to make more-informed decisions about treatment options for patients. Additionally, the reduced sample size needed for Raman spectroscopic resistance testing means that resources can be conserved. The goal is to make it easier to scale up testing in settings with limited resources.
Commercialization
The second panel in Figure 2 visualizes the system development stage. The team, led by Jürgen Popp, professor of physical chemistry at the University of Jena and scientific director of Leibniz IPHT, is working together with the University Hospital of Jena to develop diagnostic concepts and translate them into technological solutions that enable the handling of small sample volumes and cell populations to produce statistically significant and reliable measurements.
Startups in the fields of biotechnology, medical technology, or in vitro diagnostics are expected to bring these technological approaches to the market. This also involves reducing the necessary sample volume and pre-cultivation durations. Small and portable Raman microscopes, designed to measure cell characteristics essential for diagnosis, can analyze these cartridge and chip systems. A data analysis protocol has been developed and published by photonic data scientists to establish standardized procedures for processing Raman spectral data in a consistent and reliable manner6.
In a specific example, company Biophotonics Diagnostics GmbH converted this methodology into user-friendly software named RAMANMETRIX, which was designed to overcome challenges faced during the analysis of Raman spectra by using a graphical user interface, a complete workflow for the relevant data9.
The technological developments in clinical Raman spectroscopy have been oriented toward unmet clinical needs and the challenges in obtaining therapy-relevant information. Future advancements in blood diagnostics should be able to reach the patient’s bedside for direct testing and analysis. Efforts should include simple and time-saving sample handling, the development of robust and user-friendly instruments, and the creation of analysis software for the targeted evaluation of all therapy-relevant information.
It is of paramount importance for spectroscopists and clinicians to identify sample materials containing time-critical and therapy-relevant information with minimal preparation. The implementation of appropriate technological solutions in hardware and software that are currently in development will enable the use of photonic diagnostic solutions in the presence of the patient, creating new opportunities. In the future, machine learning and AI systems will be used to extract information from combined data collection approaches.
This integration will connect data from various near-patient measurement systems, enhancing the speed and efficiency of diagnostic processes. The globally unique infrastructure at the Leibniz Centre for Photonics in Infection Research (LPI) as an open-user one-stop agency is a model for how to accelerate the development of market-ready light-based diagnostic procedures and novel therapeutic approaches for the treatment of infectious diseases.
Meet the authors
Anja Silge works as a scientist at the Leibniz Institute of Photonic Technology (IPHT). She specializes in applied bio spectroscopy, enabling the straightforward analysis of patient samples in modular, easy-to-use diagnostic systems. Her objective is to find solutions that align biological and technological parameters so that specific diagnostic questions can be answered; email: [email protected].
Jürgen Popp, professor of physical chemistry at the University of Jena and scientific director of the Leibniz Institute of Photonic Technology (IPHT) has been working on using Raman spectroscopy for medical diagnoses for more than 20 years. Popp is spokesperson of the Leibniz Center for Photonics in Infection Research (LPI); email: [email protected].
Acknowledgments
Funded by the Federal Ministry of Education and Research (BMBF) Project InfectoXplore (13GW0459A), ReHwIN (13GW0432E), and SARS-CoV-2Dx (13N15745); BMBF funding program Photonics Research Germany, integrated into the Leibniz Center for Photonics in Infection Research (LPI: 13N15466, 13N15704, 13N15715). The LPI initiated by Leibniz-IPHT, Leibniz-HKI, UKJ, and FSU Jena is part of the BMBF national road map for research infrastructures. Supported by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under Germany’s Excellence Strategy — EXC 2051 — Project-ID 390713860.
References
1. J.M. Cavaillon et al. (2020). Sepsis therapies: learning from 30 years of failure of translational research to propose new leads. EMBO Mol Med, Vol. 12, No. 4, p. e10128.
2. J. Kirchhoff et al. (2018). Simple Ciprofloxacin Resistance Test and Determination of Minimal Inhibitory Concentration within 2 h Using Raman Spectroscopy. Anal Chem, Vol. 90, No. 3, pp. 1811-1818.
3. A. Ramoji et al. (2021). Leukocyte Activation Profile Assessed by Raman Spectroscopy Helps Diagnosing Infection and Sepsis. Crit Care Explor, Vol. 3, No. 5, p. e0394.
4. N. Arend et al. (2020). Detection and Differentiation of Bacterial and Fungal Infection of Neutrophils from Peripheral Blood Using Raman Spectroscopy. Anal Chem, Vol. 92, No. 15, pp. 10560-10568.
5. A. Pistiki et al. (2021). Biochemical Analysis of Leukocytes after In Vitro and In Vivo Activation with Bacterial and Fungal Pathogens Using Raman Spectroscopy. Int J Mol Sci, Vol. 22, No. 19, p. 10481.
6. S. Guo et al. (2021). Chemometric analysis in Raman spectroscopy from experimental design to machine learning-based modeling. Nat Protoc, Vol. 16, No. 12, pp. 5426-5459.
7. I.W. Schie et al. (2018). High-Throughput Screening Raman Spectroscopy Platform for Label-Free Cellomics. Anal Chem, Vol. 90, No. 3, pp. 2023-2030.
8. U.C. Schröder et al. (2013). Combined dielectrophoresis-Raman setup for the classification of pathogens recovered from the urinary tract. Anal Chem, Vol. 85, No. 22, pp. 10717-10724.
9. D. Storozhuk et al (2022). RAMANMETRIX: a delightful way to analyze Raman spectra. arXiv, arXiv:2201.07586.