A look at fluorescent protein developments reveals what’s behind them – and what the future may hold.
Hank Hogan, Contributing Editor
Thanks to ongoing efforts by researchers, fluorescent proteins continue to shine. Recent developments have taken advantage of growing knowledge to engineer brighter proteins, to produce ones that are more photostable and to create pairs that can be placed close together without drowning each other out. There also are new complementary tools that give researchers even more fluorescent choices.

Tubes of purified fluorescent proteins were photographed with UV illumination. Courtesy of Nathan C. Shaner.
Vladislav Verkhusha, an associate professor at Albert Einstein College of Medicine of Yeshiva University in Bronx, N.Y., sees a change in how new fluorescent proteins are being developed. Researchers no longer have to search for new, wild-type proteins with the desired characteristics. Instead, they can construct what they need out of what they already have.
“They can rationally design potentially any spectral or photochemical property using one of the available monomeric fluorescent proteins as a template, a starting point, for the following directed molecular evolution,” Verkhusha said.
In the past, researchers cloned proteins from marine animals, looking for those with interesting or unusual properties. Those that seemed promising were converted from their native tetrameric form of four identical units into a monomeric, or single unit, version. Researchers then enhanced the naturally present new and desirable property.
New game in town
That hunting for fluorescent proteins is no longer the only game in town, thanks to better understanding of chromophore formation pathways and improvements in high-throughput screening techniques. Together these allow researchers to dial-in a desired property.
An example of how this works can be found in a paper scheduled for publication in the October issue of Chemistry & Biology that Verkhusha co-authored. The group of researchers previously had created a mutant of the Aequorea victoria green fluorescent protein, making it form a red chromophore. In their latest work, they created blue mutants of several red fluorescent proteins, including the widely used TagRFP and mCherry. The new monomeric blue fluorescent protein mTagBFP was substantially brighter, had a faster maturation and higher pH stability than existing blue fluorescent proteins, they reported.
The better performance of the new protein, Verkhusha said, comes from the tyrosine residue in the chromophore. Previous blue fluorescent protein chromophores have contained histidine, which has a lower extinction coefficient than tyrosine.
The strategy employed in creating both the red and blue chromophores is similar and is based upon their formation pathways. Tyrosine-based red chromophores, as well as their green counterparts, are anionic, or negatively charged. During formation, they undergo two oxidations, whereas the green chromophores undergo only one. To make the red proteins blue, the researchers rationally designed site-specific mutagenesis to block the second oxidation, which caused the chromophore to acquire a form like a green fluorescent protein. At the same time, other mutations caused the chromophore to acquire a neutral, or protonated, form. The result was a blue protein, which the researchers then enhanced through random mutagenesis and selection of the most promising variants.
“The next step will be to develop monomeric fluorescent proteins that change the fluorescence colors from blue to red over time,” Verkhusha said. They’ll do this, he explained, by slowing down the rate of the second oxidation so that the protein will transition slowly from a once-oxidized blue to a twice-oxidized red. The researchers also plan to explore other possibilities based upon their knowledge of the chemical mechanism behind red chromophore formation.
A mixed picture
It isn’t only wavelength or brightness that is being improved. Nathan C. Shaner, now a postdoctoral fellow at Monterey Bay Aquarium Research Institute in Moss Landing, Calif., was the lead author of a June Nature Methods paper. At the time this research was done, Shaner was at the University of California, San Diego, in La Jolla in Roger Y. Tsien’s group, where he was involved in engineering fluorescent proteins.
In the paper, the researchers report on an assay for screening libraries of fluorescent proteins for enhanced photostability, a trait often treated as an afterthought. They used a solar simulator from Spectra-Physics of Mountain View, Calif., placing bandpass filters from Chroma Technology Corp. of Rockingham, Vt., in the optical path to provide light from 525 to 555 nm or from 548 to 588 nm at a relatively low intensity over a 10-cm2 area. With this, they illuminated bacterial colonies expressing a fluorescent protein under evaluation.
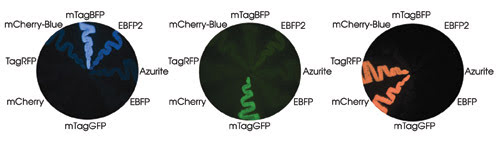
Strikes of bacteria on a petri dish are shown expressing blue mTagBFP and mCherry-Blue proteins, which have tyrosine in the chromophores. Their ancestral red TagRFP and mCherry, respectively, also are shown. Other blue proteins, including EBFP, Azurite and EBFP2, contain histidine in the chromophore. Courtesy of Vladimir Verkhusha, Albert Einstein College of Medicine.
That source was enough to photobleach the starting fluorophores of interest to half their initial intensity in as little as 10 minutes. This fast response allowed the investigators to screen libraries of up to 100,000 clones, looking for those that were more photostable than their brethren. They selected those that seemed most promising, that performed mutagenesis, and then, that screened again, repeating the process over and over.
Using this approach, they developed a red fluorescent protein variant, TagRFP-T, with nine times more photostability than its parent. In another case, they used the technique to create mOrange2, which was 25 times as photostable as its ancestor. Both fluorescent proteins, the group said, would make excellent fusion partners when expressed in mammalian cells.
This technique could be added to the other standard methods to engineer proteins that have emerged, helping to create more and better fluorescent proteins. However, Shaner noted that simply having more choices is a mixed blessing for end users.
On one hand, a greater number of choices increases the likelihood that one will work. On the other hand, ferreting out the information needed to make the correct decision can be challenging. Fortunately, Shaner and others have been involved in developing ways to better gauge the suitability of a protein.
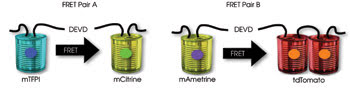
These dual FRET pairs are spectrally distinct and can be imaged in a single cell. Researchers fused the effector caspase substrate Asp-Glu-Val-Asp (DEVD) between the two fluorescent proteins of the respective FRET pairs in making a biosensor capable of signaling an increase in the cell death process-related biochemical caspase-3. Courtesy of Robert Campbell, University of Alberta.
For a more sophisticated use of fluorescent proteins, some researchers construct a FRET-based probe that enables the interaction between two fluorophores, known as the Förster or fluorescence resonance energy transfer, to track biochemical activity. Thanks to ongoing development efforts, end users have many construction examples to follow in the literature. That makes the job of creating a probe for a particular situation easier.
What is lacking, according to Shaner, is a quick way to generalize the process to make a sensor with a high dynamic range that will work with a particular cell type. “Optimization of these sensors is largely empirical and requires a lot of time and effort. This is a situation that is unlikely to improve in the near future.”
No more low-hanging fruit
The May issue of Nature Methods featured dual pairs of such FRET-based probes, developed by a team headed by Robert E. Campbell, Canada research chair in bioanalytical chemistry at the University of Alberta in Edmonton. The researchers engineered spectrally distinct fluorescent protein pairs, enabling both to be imaged at the same time. By doing so, they determined the intensity ratio of the two pairs. Such ratiometric imaging sidesteps the problems of unknown and varying output that can plague simple intensity measurements.
Building a dual FRET pair involves four fluorescent proteins and runs up against a basic problem, noted the researchers. Fluorescent proteins have excitation and emission profiles that often are close to, if not more than, 100 nm wide, if the points of one-tenth maximum intensity are considered. Consequently, placing four proteins into one intracellular locale is likely to lead to spectral bleed-through, with the excitation or emission of one affecting the others.
The group overcame this problem thanks to an unexpected find. During the development of a blue fluorescing protein, the researchers discovered a violet-excitable yellow-fluorescing variant. Through what they characterized as aggressive directed protein evolution, they turned this into a brightly fluorescing form with moderate photostability. The new fluorescent protein, mAmentine, has an emission profile very close to that of another protein, mCitrine. However, the two differ so much in excitation profiles that one can be excited in the presence of the other with little spectral crosstalk.
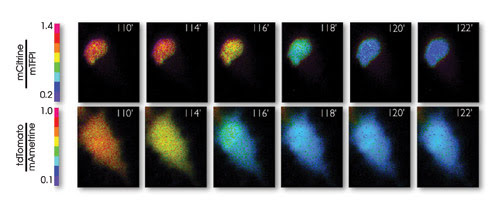
Pseudocolored ratio images of a representative staurosporine-treated HeLa cell expressing dual caspase biosensors that monitor cell death in the nucleus and cytoplasm. The FRET change resulting from activation of caspase-3 occurs in the cytoplasm approximately 2 min before the change occurs in the nucleus. Time shown is in minutes; color bar represents fluorescence ratio. Reprinted from Nature Methods with permission of the researchers.
The researchers used that separation in assembling their two FRET pairs as part of a biosensor for caspase-3, one of a family of biochemicals that play essential roles in programmed cell death, necrosis and inflammation. With these sensors, they followed caspase-3 during cell death, detecting an increase of the biochemical in the cytoplasm with one FRET pair and in the nucleus with the other.
Campbell said that fluorescent proteins are definitely improving with time. But he also noted that most of the low-hanging fruit has already been picked.
Future objectives for protein engineers should include improving photostability, shifting fluorescence to the near-infrared and overcoming other large hurdles. “One of the major challenges for the future is to use these improved fluorescent proteins to make large-dynamic-range sensors for a variety of key biological events,” he said.
Expanding toolboxes via insects
Of course, fluorescent protein research is not confined to academe. Invitrogen Corp. of Carlsbad, Calif., for example, offers its own line of fluorescent proteins as well as related and complementary products. Organic dyes once were an exclusive focus, but the company has branched out over the past few years. For instance, besides fluorescent proteins, it now has quantum dot-based products that exploit the photostability and narrow spectral emission of these semiconductor nanocrystals.
Magnus Persmark, the company’s senior product manager of labeling and detection, points to two recent developments as examples of a trend toward broadening the fluorescent toolbox. Both are based upon combinations of technologies found within the company. Each uses a fluorescent probe to stain the parts of the cell of interest, thereby highlighting them. This approach is particularly important in industrial settings, where rapid results are required and it is not possible to wait for the development of a stable transfected cell line.
The first addition to the toolbox is based on BacMam technology, which uses an insect cell virus to deliver a genetic payload to a mammalian cell. As a result of its insect origins, the virus can penetrate the cell in question but cannot replicate. Because of that, BacMam virions can be used on primary and stem cells, which can be more representative of cells within a living organism than cell lines. The transiently expressed fluorescent protein genes are tagged with peptides so that they stain specific subcellular organelles a given color.
“Some of the products we have come up with result in mitochondrial green, red or orange staining of living cells,” Persmark said. Other stains, he added, highlight the Golgi apparatus, plasma membrane or another cellular structure.
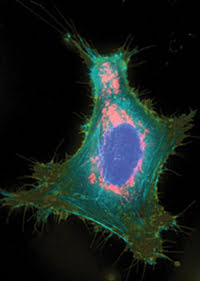
A HeLa cell is expressing the spectrally distinct fluorescent proteins mAmetrine (membrane), mTFP1 (actin), EYFP (nucleus) and tdTomato (mitochondria) in targeted fusion constructs. Reprinted from Nature Methods with permission of the researchers.
He reported that customers use this technology when doing colocalization studies, drug discovery or other cell-based research. They may choose to do so because of the technology’s relative simplicity. For example, it is possible to modulate the expression of the transiently transfected protein by simply altering the dose of the BacMam reagent, something that cannot be done easily with a stable cell line containing a genetically encoded protein.
The company also offers a line of quantum dot- and organic dye-labeled antibodies, which can be complementary to fluorescent proteins. These antibody conjugates detect antigens in fixed cells and offer some unique characteristics. For example, a quantum dot’s resistance to photobleaching and ability to multiplex fluorescent signals can be useful.
However, given their ability to be introduced into cells using genetic techniques, it is likely that fluorescent proteins will continue to be the technology of choice for many researchers engaged in live cell studies. Being produced with a cell rather than being introduced from the outside avoids a host of issues. It also permits analysis of a cell-based model that is in as close to a normal environment as possible.
With regard to the development of fluorescent proteins, Persmark noted that, as understanding of the chromophores grows, researchers are indeed better able to direct development to produce a protein with desired characteristics. But he does not see such a capability as being the primary source for future major advances.
Instead, he cites groundbreaking live cell sensors developed by Tsien, Atsushi Miyawaki of Japan’s Riken Institute and co-workers as examples of the direction of future developments.
“Where the breakthroughs are going to be is probably more in terms of the applications than in terms of the tools, the fluorescent probes, themselves,” Persmark said.