Surface plasmon resonance technology provides high information content in proteomics.
Alan McWhirter, Biacore AB
Proteins are the worker bees
of the human body. They traverse cell membranes to receive and propagate signals,
transport oxygen to every cell in the body, regulate the expression of genes and
impart structure and strength to tissues. The signaling networks that transmit information
from stimulus to biological function, from the environment to the organism, from
cell to cell within tissues and within cells themselves are all regulated by proteins.
Studying proteins, therefore, is critical to understanding how specific ones contribute
to specific biological functions.
One method uses surface plasmon resonance (SPR),
a natural phenomenon that occurs when polarized light strikes an electrically conducting
gold layer.
Until now, SPR-based biosensors have
delivered high-quality data on specific interactions. But with a bewildering number
of novel proteins at hand since the completion of the human genome project, the
proteomics community needs a high-quality, high-throughput method of studying their
interactions.
Additionally, applications such as
antibody screening, drug development and even online quality control/safety testing
during food production could see the practical and economic benefits from a method
such as an SPR array, which offers both high sample throughput and a large amount
of information on interactions.
Watching interactions
Before proteins can influence function, they must
communicate. To communicate, they must interact with other proteins or with other
large biomolecules or small molecules such as vitamins or nucleotides. While engaged
with an interacting partner, proteins drive (or participate in the regulation of)
a function; when they dissociate, the function ceases.
Clearly then, quantitative data that
describe how proteins interact — the rates of association and dissociation,
called the interaction profile — will enable informed judgments about the
contribution that individual proteins make to fine-tuning the complicated and extensive
protein networks that regulate biological events.
For example, many proteins in intracellular
signaling networks are active (capable of binding a partner) only when they are
phosphorylated on specific amino acid residues. Phosphorylation status is regulated
by two sets of proteins, called kinases and phosphatases, which phosphorylate and
dephosphorylate proteins, respectively. The cellular activity (e.g., motility, DNA
replication, protein synthesis, apoptosis) regulated by the pathway in which these
proteins participate will be influenced by the phosphorylation status of proteins
throughout the signaling network — a fine kinetic balancing act of coordinated
associations and dissociations.
In addition, pharmaceutical companies
determine whether a compound continues in drug development processes based on interaction
profiles with targets in relation to their intended function. An ideal anticancer
drug, for example, is likely to be characterized by rapid association and slow dissociation
from its protein target. A dental anesthetic, on the other hand, should dissociate
from its target rapidly so that the patient can quickly return to normal after a
filling rather than remaining anesthetized for a week.
Charting protein networks
Proteomics is the collective term for the many
investigative strategies delivering data that document the proteins that a genome
encodes (collectively known as a proteome) and those formed after posttranslational
modifications.
Listing a few descriptors of a proteome,
such as molecular weight and domain properties, is rather like compiling the telephone
directory of a town: Until the information is used — and people start calling
each other — it is of little intrinsic value. Similarly, the repository of
data comprising the proteome is somewhat lifeless until we use it to understand
what all these thousands of proteins actually do and how they relate to one another.
Thus, instruments that can rapidly
profile hundreds of protein interactions are attractive to investigators who would
like to use the vast repository of data from proteomics initiatives to solve real
problems. SPR-based arrays can offer large amounts of information on protein interactions
in a high-throughput manner.
One major benefit of SPR over technologies
such as ELISA or affinity chromatography is that it can provide high-resolution
kinetics in real time over the course of an interaction. This provides a comprehensive
and detailed profile of association and dissociation, imparting information about
protein function far beyond that which can be inferred from end-point assays.
For example, by studying the rates
of association or dissociation, a scientist could deconstruct a protein complex
in terms of recognition or stability, forming a basis for qualified proposals of
an interaction model. Furthermore, as the status of an interaction is followed
according to changes in mass close to a sensor surface (i.e., as a molecular complex
forms and dissociates), there is no requirement to label any of the interacting
partners.
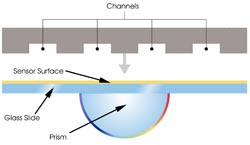
Figure 1. In prism-based surface plasmon resonance systems, a block
engraved with channels is pressed against a sensor surface to create a flow cell,
the site of protein interactions on which one partner is immobilized and over which
the other passes in solution.
SPR-based protein arrays are designed
according to the type of information required. The Biacore A100 uses prism-based
SPR (Figure 1), which occurs when polarized light, under conditions of total internal
reflection, strikes an electrically conducting gold layer at the interface between
media of different refractive index; the glass of the sensor surface covered with
a thin layer of gold (high refractive index) and a buffer flowing over the sensor
surface (low refractive index).
A wedge of polarized light, covering
a range of incident angles, is directed toward the glass face of the sensor surface
and reflected toward a detector. When the light strikes the glass, it generates
an electric field intensity known as an evanescent wave, which interacts with
and is absorbed by free electron clouds in the gold layer, producing electron charge
density waves called plasmons and causing a reduction in the intensity of the
reflected light.
Interactions can be detected because
the resonance angle at which this intensity minimum occurs is a function of the
refractive index and, hence, the molecular mass of the complex adjacent to the gold
layer on the sensor surface (Figure 2).
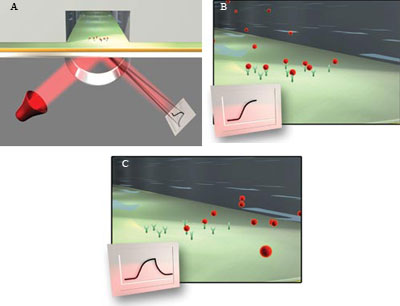
Figure 2. As molecules are immobilized on a sensor surface, the refractive
index at the interface between the surface and a solution flowing over it changes,
altering the angle at which reduced-intensity polarized light reflects from the
supporting glass plane. The change in angle, caused by binding or dissociation of
molecules from the sensor surface, is proportional to the mass of bound material
and is recorded in a sensorgram (A). When sample in solution passes over the sensor
surface, the sensorgram shows an increasing response if the molecules interact.
The response remains constant after the interaction reaches equilibrium (B). When
sample is replaced by buffer, the response decreases as the interaction partners
dissociate (C).
Simultaneous profiling
The Flexchip system allows simultaneous profiling
of up to 400 protein interactions and uses an alternate arrangement called grating-coupled
SPR (Figure 3), where incident polarized light from above strikes the entire functional
face of a finely grated sensor surface. The small coupling angle of the incident
light is conducive to multiple imaging and thus well suited to screening applications
requiring simultaneous interrogation of a large number of interactions.
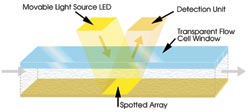
Figure 3. In systems based on grating-coupled
SPR, incident light from above directly strikes the entire array, simultaneously
generating data from as many as 400 interactions.
In instruments from Biacore AB of Uppsala,
Sweden, biomolecular interactions are studied in flow cells on a sensor surface.
In this setup, the researcher injects interacting partners in solution into flow
cells that are formed when a microfluidic flow system is brought into contact with
a sensor surface on which other interacting partners have been immobilized.
Hydrodynamic addressing is performed
using a single flow cell in which multiple targets may be immobilized on detection
spots (the site of interaction), allowing simultaneous analysis of interactions.
By adjusting the relative flow at the two inlets — one for the immobilized
partner and the other for the buffer — liquid can be directed to one or another
of the addressable detection spots (Figure 4).
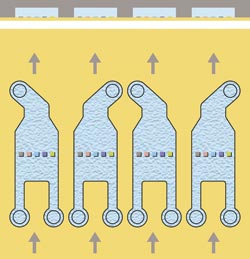
Figure 4. Each flow cell has two inlets and one outlet, allowing the spots to be addressed
separately. No target is immobilized on the central spot, which is used as a reference.
The design of the flow cell and the simultaneous flow of sample solution over all
five spots are essential components in small-molecule applications, providing simultaneous
binding analysis to four target spots with in-line referencing.
The A100, for example, has four parallel
independent hydrodynamic addressing flow cells that can accommodate up to five proteins
immobilized in each. For maximum sample throughput, identical immobilizations can
be performed in each cell, allowing analysis of four samples in parallel during
each analysis cycle. In assays where information output per sample is more important,
up to 20 interactants can be immobilized in the four flow cells, and one sample
per cycle is injected in parallel over all flow cells.
Rapid screening
Productivity in biotherapeutic development may
benefit from access to a high-throughput array system that delivers kinetic data.
For example, the development of monoclonal antibodies is a complex and time-consuming
process, involving the generation, maintenance and screening of thousands of hybridoma
clones — the cellular “factories” in which these antibodies are
produced. Early identification of the clones that produce the best candidate monoclonal
antibodies is a critical step in development. Rapid screening efficiently enables
selection of candidates with the required kinetic profiles, discriminating between
equal-affinity antibodies based on kinetic properties that are crucial for clinical
success.
In addition, this array system can
help in testing how likely newly developed drugs and vaccines are to elicit an immune
response. This is important because the human body may identify even the most carefully
designed and constructed drug as a foreign body, causing an unwanted antibody response.
SPR-based protein arrays can be used to characterize serum antibody responses by
detecting potential clinically relevant low- and medium-affinity antibodies, producing
data on isotype, subclass specificity and kinetics from a single system using low
quantities of sera.
Flexchip uses a single-pass multispot
flow cell, a single broad channel through which sample is injected. The sample interacts
simultaneously with up to 400 spots on a single array (Figure 5). As light reflects
from the sensor surface, the system’s software resolves the data from the
individual spots into hundreds of interaction profiles.

Figure 5. The design of this flow cell is a single broad channel
through which sample is injected, interacting simultaneously with all spots on an
array. After the sensor surface is spotted according to user specifications, a gasketed
window with an inlet and an outlet valve is positioned and hermetically sealed over
the array to form the flow cell, which is inserted into the Flexchip apparatus.
Researchers can study interactions
between proteins using a peptide from one of the interacting partners rather than
from the whole protein. This type of array could, for example, identify peptides
with the highest binding activity by looking for overlapping peptides on the sequence
of one interacting partner. The array may then be further applied in an alanine
scan, in which single amino acid residues in a peptide are replaced by alanine,
to precisely identify the amino acid residues necessary for the interaction.
The array also could be used to define
how transcription factors bind to DNA by comparing interactions with wild-type DNA
oligomers with those containing mutations within the consensus sequence. Electrostatic
interactions have been reported to influence the association of transcription factors
to DNA, and this may be mimicked in vitro by increasing the ionic strength of the
running buffer, a condition that tends to favor specific over nonspecific interactions.
Interactions may thus be followed at different ionic strengths over a series
of runs and the interaction profiles compared.
Meet the author
Alan McWhirter is an academic sector specialist in market communications at Biacore AB in Uppsala, Sweden; e-mail: [email protected]m.