A hybrid system creates an analytical tool that enables the discovery of hidden signatures in cancer cells.
CHIEN-SHENG LIAO, WELDON SCHOOL OF BIOMEDICAL ENGINEERING, PURDUE UNIVERSITY, AND JI-XIN CHENG, WELDON SCHOOL OF BIOMEDICAL ENGINEERING, PURDUE UNIVERSITY, AND THE PHOTONICS CENTER, BOSTON UNIVERSITY
Most of our knowledge about the chemical contents in cancer cells or cancerous tissue is obtained from in vitro analysis of fixed cells and tissues or homogenates. These in vitro assays identify the presence of molecules, but do not provide information about their spatial distribution or temporal dynamics. A detailed mapping of molecules in live cells or intact tissues would pave the way for disease-specific drug developments or better marker-based diagnosis.
Raman scattering, an inelastic scattering process first reported in the 1920s by C.V. Raman and others, enables label-free, quantitative analysis of molecules in cells and tissues. In this inelastic process, excitation photons are scattered by the molecules and part of the energy of incident photons induces molecular vibrations (Ω). The scattered photons, which have less energy, shift to the red end of the spectrum and generate new colors from the original photons (Figure 1). The spectrum of these scattered photons, namely, the Raman spectrum, exhibits information about the vibrational energy that can be used to identify chemical bonding and modes of vibrations in molecules. Essentially, Raman spectroscopy provides the “fingerprint” information that resolves the molecular composition in cells or tissues. Confocal Raman microscopy further enables visualization of the spatial location of molecules and their interactions with environments with a typical acquisition time of a few seconds per pixel.
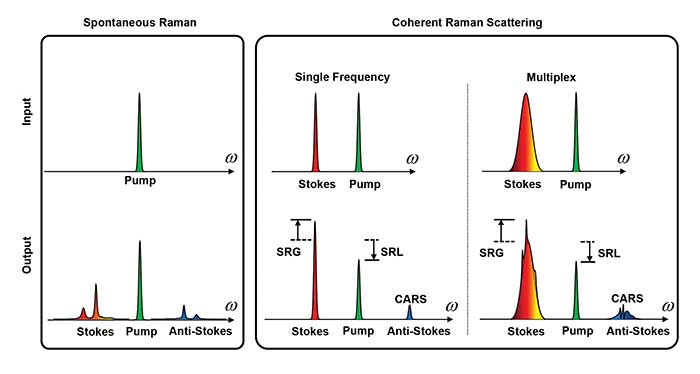
Figure 1. Spontaneous Raman spectroscopy (left) covers the entire molecular vibration and allows extraction of subtle compositional features by multivariate analysis. Coherent Raman microscopy (right) provides a speed advantage by focusing energy into either a single Raman band or into a defined spectral window for target molecules. CARS: coherent anti-Stokes Raman scattering; SRL: stimulated Raman loss; SRG: stimulted Raman gain. Courtesy of Ji-Xen Cheng. Adapted with permission from “Vibrational spectroscopic imaging of living systems: An emerging platform for biology and medicine,” Science. Copyright 2015, American Association for the Advancement of Science.
As an advanced Raman technique, coherent Raman scattering microscopy based on coherent anti-Stokes Raman scattering (CARS) and stimulated Raman scattering (SRS) has been developed to speed up the acquisition of molecular vibrational signals1. By focusing on a single Raman band, the imaging speed has reached video rate in both CARS2 and SRS microscopy3.
CARS involves three laser fields, namely a pump field at frequency of ωp, a Stokes field at ωS , and a probe field at ωpr. When the energy difference of the pump and Stokes beams (ωp−ωS) matches the frequency of a molecular vibration mode (Ω), the Raman resonance occurs. This resonance is then probed by the third field at ωpr, generating an anti-Stokes field at (ωp−ωS+ωpr) (Figure 1).
SRS is a dissipative process that occurs simultaneously with CARS. When the beating frequency (ωp−ωS) is tuned to excite a molecular vibration, the energy difference between ωp and ωS pumps the molecule from a ground state to a vibrationally excited state. The laser field manifests itself as a weak decrease in the pump beam intensity, called stimulated Raman loss, and a simultaneous increase in the Stokes beam intensity, called stimulated Raman gain (Figure 1). Because the Raman gain or loss signal appears at the same frequency as one of the incident beams, optical modulation and demodulation are needed to extract the SRS signal.
Coherent Raman and spontaneous Raman are complementary to each other. Spontaneous Raman spectroscopy covers the entire molecular vibration and allows extraction of subtle compositional features by multivariate analysis. Coherent Raman microscopy provides a speed advantage over spontaneous Raman by focusing energy into either a single Raman band or into a defined spectral window for target molecules. The technology has led to recent advances in cancer research.
Revealing hidden signatures
Spontaneous Raman spectroscopy and coherent Raman microscopy, when integrated, can be a powerful analytical tool that enables discovery of hidden signatures in cancer cells or tissues. Using this hybrid Raman system, the Cheng group discovered cholesteryl ester as a metabolic marker of aggressive prostate cancer4. More recently, Cheng and coworkers found lipid desaturation as a metabolic marker and therapeutic target of ovarian cancer stem cells (Figure 2)5.
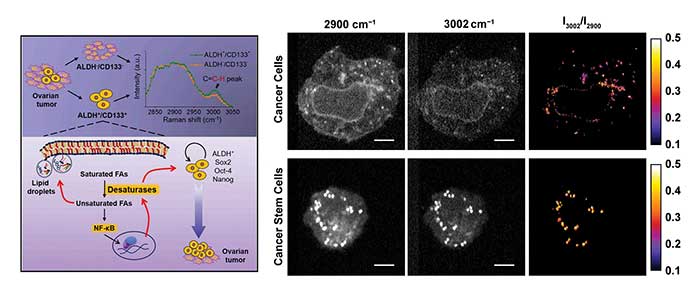
Figure 2. A hybrid system using both spontaneous Raman spectroscopy and coherent Raman microscopy can uncover hidden signatures in cancer cells. SRS images at 3002 cm−1 of vibration of = C-H bonds are mostly from unsaturated lipids. Images at 2900 cm−1 of C-H stretch vibrations are from all the lipids. Courtesy of Ji-Xen Cheng. Adapted with permission from “Lipid desaturation is a metabolic marker and therapeutic target of ovarian cancer stem cells,” Cell Stem Cell. Copyright 2016, Elsevier Inc.
Cancer stem cells represent a small subpopulation of cancer cells that have the properties of self-renewal and initiation of tumors in vivo. Understanding the characteristics and targeting the vulnerabilities of cancer stem cells will enable the development of therapies capable of overcoming tumor relapse and metastasis. Raman-based analytical tools provide exclusive advantages for the characterization of metabolic activity in cancer stem cells in a label-free manner. With the aid of hyperspectral SRS microscopy covering the feature vibrational peak of =C-H bonds that are mostly in unsaturated lipids, Cheng and coworkers recently discovered significantly increased levels of unsaturated lipids in ovarian cancer stem cells compared to noncancer stem cells5. Spontaneous Raman spectroscopy focusing on the lipid droplets in cancer stem cells confirmed the finding based on the Raman peak at 1264 cm−1 contributed by the plane-bending motion of =C-H and the peak at 1660 cm−1 contributed by the stretching vibration of C = C. In addition, inhibition of lipid desaturases showed effective elimination of ovarian cancer stem cells, suppression of the sphere formation in vitro and the block of tumor initiation capacity in vivo. The unsaturation level as the marker representing a targetable metabolic vulnerability of recalcitrant stem cells heralds the possibility of improving the outcome of existing cancer therapies.
Chemical histology
Raman signals are used to identify cancer cells from healthy tissue based on their inherent molecular compositions. Conventionally, histopathological assessment of in vitro hematoxylin and eosin (H&E)-stained tissue is the gold standard to establish diagnosis of malignancy. The sectioning and labeling processes are typically labor-intensive and time-consuming. This difficulty can be addressed by coherent Raman imaging, which can resolve spatial distribution of cancer cells. Taking brain tumor as one example, healthy regions of brain including lipid-rich white matter and cortex exhibit Raman signals at 2850 cm−1 of CH2 stretching mode and 2940 cm−1 of CH3 stretching mode, while protein-rich tumor shows signals mostly at 2940 cm−1.
By focusing on these two Raman bands, the Sunney Xie group used two-color SRS microscopy to detect the glioma in human xenograft mice and neurosurgical patients6 (Figure 3). The image results were correlated with the interpretation of H&E slides by a surgical pathologist. The brain tumor infiltration was quantified by alterations in tissue cellularity, axonal density and protein/lipid ratio.
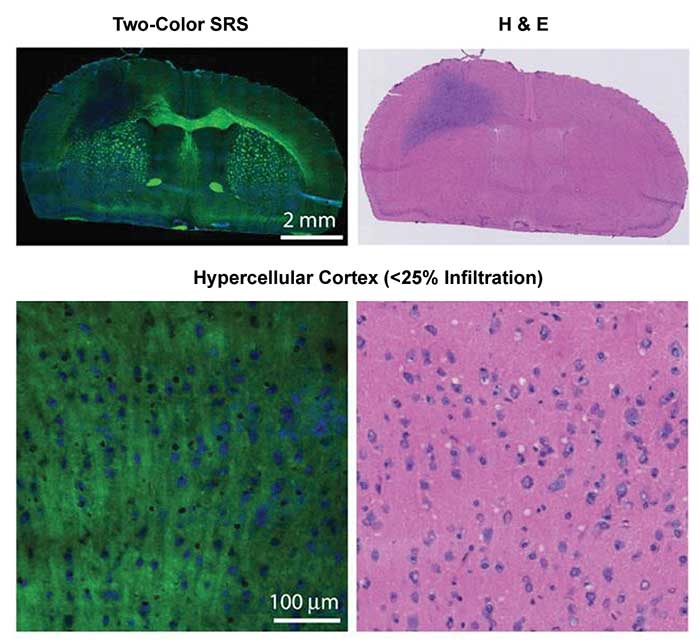
Figure 3. Healthy regions of brain including lipid-rich white matter and cortex exhibit Raman signals at 2850 cm−1 of CH2 stretching mode and 2940 cm−1 of CH3 stretching mode, while protein-rich tumor shows signals mostly at 2940 cm−1. Two-color SRS (left) microscopy detected glioma in human brain cancerous tissues that correlated with the interpretation of hematoxylin and eosin (H&E) slides (right) by a surgical pathologist. Adapted with permission from Dr. Sunney Xie et al., Rapid, label-free detection of brain tumors with stimulated Raman scattering microscopy, Sci Transl Med. Copyright 2013 American Association for the Advancement of Science.
Eric Potma and coworkers used SRS microscopy focusing at 2945 cm−1 to identify pathological features of squamous cell carcinoma in skin cancer7. This Raman band exhibiting the contrast of protein density distinguished the cell nucleus from the cell cytoplasm, and the results showed a close correlation with the diagnostic features identified by H&E histopathological contrast.
In the same spectral window of CH2 and CH3 stretching modes, the Stephen Boppart group developed interferometric CARS microscopy to collect a vibrational spectrum with accurate Raman line shape at each pixel of the imaged tissue specimen and applied it to a preclinical rat breast cancer model8. This approach differentiated cancer cells from normal tissue sections with greater than 99 percent accuracy.
Cancer detection in the clinic
Coherent Raman microscopy has shown the ability of generating histopathological images that resemble H&E-stained histologic result in vitro for many diseases. The label-free characteristic of these Raman-based diagnostic tools also holds the potential for intraoperative histological assessment, which can significantly impact patients’ survival rate and quality of life. Taking brain cancer as one example, the inability to visualize invasive brain cancers usually leads to incomplete surgical resections, which negatively affects survival rates. Currently, more than 85 percent of glioblastoma multiforme recurrences occur at the resection cavity margin. On the other hand, the removal of brain tissue that does not contain cancer cells can result in neurological deficit, which can cause impaired cognition, memory and vision. Therefore, precise mapping of residual cancer in the cavity can assist complete resection of the tumor and conserve the healthy tissue. In the case of breast cancer, off-site histology is usually performed after breast-conserving therapy to determine whether the excised tumor was surrounded by a sufficient amount of normal tissue. If a positive margin is identified, a second surgical operation is needed to reduce the rate of cancer recurrence. Currently the re-operation rate for breast cancer surgery is 30 to 40 percent.
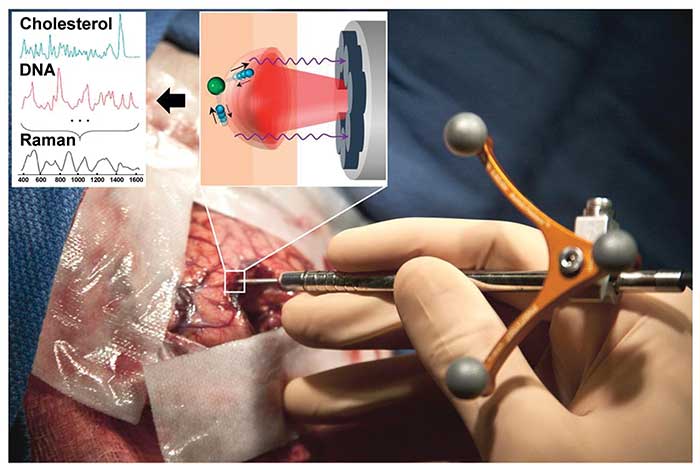
Figure 4. This handheld Raman device is used to distinguish cancerous tissue from normal brain tissue. Adapted with permission from Dr. Frederic Leblond et al., Intraoperative brain cancer detection with Raman spectroscopy in humans, Sci Transl Med. Copyright 2015 American Association for the Advancement of Science.
For brain and breast cancer surgery, technologies enabling intraoperative detection of residual cancer cells will significantly reduce the rate of re-operation and improve survival rates. The minimized resection of healthy tissue also can improve a patient’s quality of life.
Along this line, Anita Mahadevan-Jansen’s group quantified the increased relative protein contributions in breast cancerous tissues by spatially offset Raman spectroscopy, which measures Raman spectra at offset positions from the light source in order to extract information from deep tissues. The in vitro results showed potential for classification of positive and negative tumor margins ~2 mm below the surface9. Petrecca, Leblond and coworkers developed a handheld contact Raman spectroscopic probe for detection of cancer cells in the human brain intraoperatively10. This handheld spontaneous Raman spectroscopic tool, collecting one vibrational spectrum from a 0.5-mm-diameter tissue area with a depth sampling up to ~1 mm and a total acquisition time below 1 second, detected grade 2 to 4 gliomas in real time during a human brain cancer surgery. With the sensitivity and specificity of higher than 90 percent, this tool distinguished cancer-cell-invaded brain from normal brain (Figure 4).
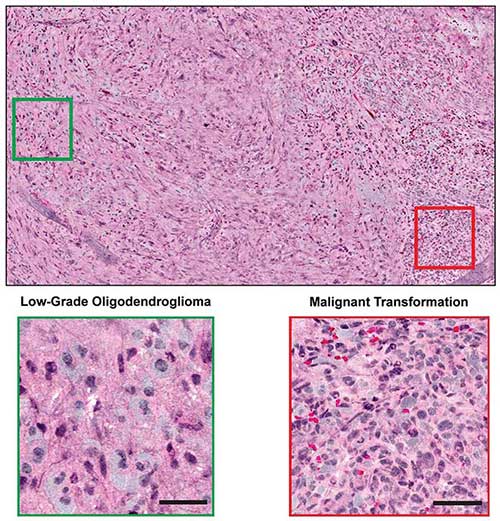
Figure 5. Combined with supervised machine learning, stimulated Raman histology by intraoperative SRS microscope effectively differentiated among diagnostic classes of brain tumors with 90 percent accuracy in neurosurgical patients. Courtesy of Dr. Daniel A. Orringer.
Orringer and colleagues recently applied a fib11. The image results showed remarkable concordance with conventional histology for predicting diagnosis and high accuracy exceeding 92 percent (Figure 5).
Combined with supervised machine learning, the quantified stimulated Raman histology image effectively differentiated among diagnostic classes of brain tumors with 90 percent accuracy in neurosurgical patients.
Meet the authors
Chien-Sheng Liao is a research assistant at the Weldon School of Biomedical Engineering, Purdue University; email: [email protected]. Ji-Xin Cheng is a professor at the Weldon School of Biomedical Engineering, Purdue University, and at the Photonics Center, Boston University; email: [email protected].
References
1. J.X. Cheng and X.S. Xie (2015). Vibrational spectroscopic imaging of living systems: An emerging platform for biology and medicine. Science, Vol. 350, aaa8870.
2. C.L. Evans et al. (2005). Chemical imaging of tissue in vivo with video-rate coherent anti-Stokes Raman scattering microscopy. Proc Natl Acad Sci USA, Vol. 102, p. 16807.
3. B.G. Saar et al. (2010). Video-rate molecular imaging in vivo with stimulated Raman scattering. Science, Vol. 330, p. 1368.
4. S.H. Yue et al. (2014). Cholesteryl ester accumulation induced by PTEN loss and PI3K/AKT activation underlies human prostate cancer aggressiveness. Cell Metab, Vol. 19, p. 393.
5. J. Li et al. (2017). Lipid desaturation is a metabolic marker and therapeutic target of ovarian cancer stem cells. Cell Stem Cell, in press, DOI: http://dx.doi.org/10.1016/j.stem.2016.11.004.
6. M.B. Ji et al. (2013). Rapid, label-free detection of brain tumors with stimulated Raman scattering microscopy. Sci Transl Med, 5, 201ra119.
7. R. Mittal et al. (2013). Evaluation of stimulated Raman scattering microscopy for identifying squamous cell carcinoma in human skin. Lasers Surg Med, Vol. 45, p. 496.
8. P.D. Chowdary et al. (2010). Molecular histopathology by spectrally reconstructed nonlinear interferometric vibrational imaging. Cancer Res, Vol. 70, p. 9562.
9. M.D. Keller et al. (2011). Development of a spatially offset Raman spectroscopy probe for breast tumor surgical margin evaluation. J Biomed Opt, Vol. 16, p. 077006.
10. M. Jermyn et al. (2015). Intraoperative brain cancer detection with Raman spectroscopy in humans. Sci Transl Med, Vol. 7, 274ra19.
11. D. A. Orringer et al. (2017). Rapid intraoperative histology of unprocessed surgical specimens via fibre-laser-based stimulated Raman scattering microscopy. Nat Biomed Eng 1, article 0027.