Variations on structured illumination microscopy (SIM) provide a way to watch dynamic biological processes inside living cells with unprecedented clarity.
"These methods set a new standard for how far you can push the speed and noninvasiveness of superresolution imaging," said Nobel laureate Dr. Eric Betzig. "This will bring superresolution to live-cell imaging for real."
A group leader at the Howard Hughes Medical Institute's Janelia Research Campus, Betzig was one of three researchers to win the 2014 Nobel Prize in chemistry for his role in developing the first superresolution optical microscopy techniques in the late 1990s.
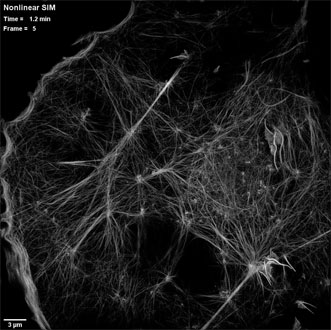
An actin cytoskeleton as seen by patterned photoactivation nonlinear structured illumination microscopy (SIM), in a COS-7 cell at 37 °C expressing the fluroescent tag Skylan-NS-Lifeact. Courtesy of the Betzig Lab, HHMI/Janelia Research Campus.
Superresolution microscopy produces images with spatial resolution beyond the diffraction limit of light, offering extraordinary visual detail of structures inside cells. Until now, leading superresolution methods have been impractical for use with living tissue because the light energies involved are destructive.
Techniques such as stimulated emission depletion operate by activating fluorescent tags in cells with one light pulse, then deactivating most of them with another. After exposure to the deactivating light, only molecules at the darkest regions of the light wave continue to fluoresce. These provide higher frequency information and sharpen the resulting image.
"The problem with this approach is that you first turn on all the molecules, then you immediately turn off almost all the molecules," Betzig said. "The molecules you've turned off don't contribute anything to the image, but you've just fried them twice. You're stressing the molecules, and it takes a lot of time, which you don't have, because the cell is moving."
The solution was simple, Betzig said: "Don't turn on all of the molecules. There's no need to do that."
One of the new techniques, called patterned photoactivation nonlinear SIM, begins by switching on just a subset of fluorescent labels in a sample with a pattern of light. Another pattern is then used to deactivate molecules, and additional information is read out of their deactivation. The process is repeated 25 times in about one-third of a second to build an image with 62-nm resolution — better than standard SIM and a three-fold improvement over the diffraction limit.

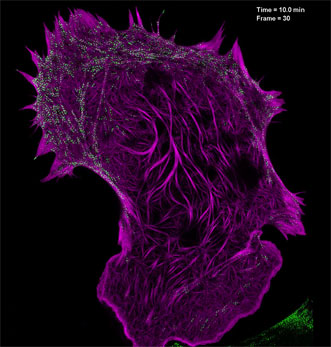
Filamentous actin (mApple-F-tractin, purple) interacts with myosin IIA bipolar head groups (EGFP, myosin IIA, green) in this image produced using high-numerical-aperature structured illumination microscopy. Courtesy of the Betzig Lab, HHMI/Janelia Research Campus.
Because the technique uses low-intensity light, it can be used to generate sequential images of cells without damaging them.
Betzig and colleagues used patterned photoactivation nonlinear SIM to produce videos showing structural proteins break down and reassemble themselves as cells move and change shape, as well as the dynamics of tiny pits on cell surfaces called caveolae.
The team also developed a way to boost the spatial resolution of SIM to 84 nm using a commercially available microscope objective with an ultrahigh numerical aperture. The aperture restricts light exposure to a very small fraction of a sample, limiting damage to cells and fluorescent molecules. The method also can be used to image multiple colors at the same time, so scientists can simultaneously track several different proteins.
Using the high-numerical-aperture approach, Betzig's team watched the movements and interactions of several structural proteins during the formation of focal adhesions, which are physical links between the interior and exterior of a cell.
They also followed the growth and internalization of clathrin-coated pits, structures that facilitate the intake of molecules from outside the cell. Their quantitative analysis answered several questions about the pits' distribution and the relationship between pit size and lifespan that could not be addressed with previous imaging methods.
Combining the high-numerical-aperture approach with patterned photoactivatable nonlinear SIM allowed the researchers to follow two proteins at a time with higher resolution than the high-numerical-aperture approach alone.
The research was published in Science (doi: 10.1126/science.aab3500).