Building on earlier superresolution methods, a new microscopy technique provides nanoscale spatial information on individual molecules while also revealing their spectra.
SR-STORM, or spectrally resolved stochastic optical reconstruction microscopy, could enable high-resolution imaging of multiple components and local chemical environments, such as pH variations, inside a cell.
"We measure both the position and spectrum of each individual molecule, plotting its superresolved spatial position in two dimensions and coloring each molecule according to its spectral position," said Ke Xu, a faculty scientist at Lawrence Berkeley National Laboratory. "So in that sense, it's true-color superresolution microscopy, which is the first of its kind."
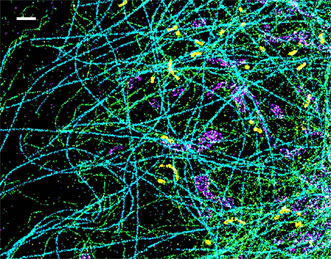
A spectrally resolved superresolution microscope image of four subcellular targets labeled by four far-red dyes at 10 nm spectral separation. Color is used to indicate the measured fluorescence emission position of each single molecule. (Scale bar: 1 μm.) Courtesy of Ke Xu/Berkeley Lab.
SR-STORM also allows high throughput, able to deliver spatial and spectral information for millions of single molecules with approximately 10-nm resolution in about five minutes. Conventional scanning-based techniques take several minutes to create a single image comprising tens of molecules.
Xu built on work he did as a postdoctoral researcher at Harvard University with professor Xiaowei Zhuang, who invented STORM, a superresolution microscopy method based on single-molecule imaging and photoswitching.
Current superresolution microscopy techniques do not deliver spectral information, which is useful for scientists to understand the behavior of individual molecules, as well as to enable high-quality multicolor imaging of multiple targets.
"So we constructed a dual-objective system but dispersed the single-molecule image collected by one objective lens into spectrum while keeping the other image for single-molecule localization," Xu said. "Now we are simultaneously accumulating the spectrum of the single molecules and also their position, so we solved the conundrum."
Using the device, Xu and colleagues showed that actin, a key component of the cytoskeleton or backbone of the cell, has a different structure in axons than in dendrites, two parts of a neuron.
Next they dyed the sample with 14 different dyes in a narrow emission window and excited and photoswitched the molecules with one laser. While the spectra of the 14 dyes overlapped heavily, the researchers found that the spectra of the individual molecules were surprisingly different and thus readily identifiable.
Using four dyes to label four different subcellular structures, such as mitochondria and microtubules, the researchers distinguished molecules of different dyes based on their spectral mean alone, and each subcellular structure was a distinct color.
"The applications are mostly in fundamental research and cell biology at this point, but hopefully it will lead to medical applications," Xu said. "This gives us new opportunities to look at cell structures, how they're built up and whether there's any degradation of those structures in diseases."
The research was published in Nature Methods (doi: 10.1038/nmeth.3528).
For more information, visit www.lbl.gov.