A new technique skips traditional histology steps to speed up diagnosis and allow for views that are not possible with conventional sectioning.
FARZAD FEREIDOUNI, AUSTIN TODD AND RICHARD LEVENSON, UC DAVIS
Even in this era of molecular medicine, conventional light microscopy remains the principal contributor to cancer diagnostics and also serves to guide surgical procedures, monitor disease activity and follow therapy responses over time. Although pathologists don’t like to admit it, the optical principles that underlie their microscopes date back, largely unchanged, to the 18th century. Moreover, for these microscopes to be useful, properly prepared tissue samples mounted onto glass slides are a prerequisite, and the methodology for the necessary histological processing has itself not changed significantly since the end of the 19th century. Both aspects of tissue diagnostics are due for a refresh.
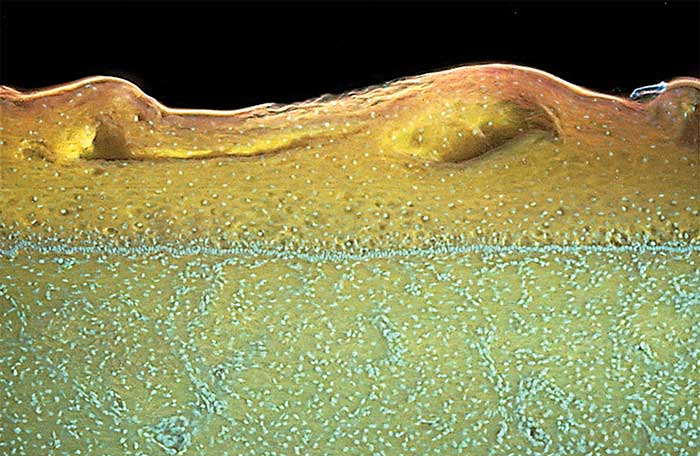
Figure 1. Squamous epithelium of the cervix above a layer of connective tissue and blood vessels. Courtesy of Richard Levenson.
Because typical light microscopes capture light that is transmitted through a specimen, for best results, the latter must be cut into very thin slices. If the sample is too thick, transillumination through multiple cell layers results in unacceptably blurred and uninterpretable images. For traditional microscopes to generate sharp, high-contrast results, tissues need to be sectioned to around 4 to 6 microns in thickness, somewhat less than that of a typical cell nucleus. Making such thin slices is routine — in well-resourced settings. But the process involves multiple steps including fixation, dehydration, paraffin-embedding, microtome sectioning, mounting, solvent treatment, staining, more solvents, and, finally, coverslipping.
The whole process can take overnight, and diagnoses are then not available until at least the day after the biopsy or surgical procedure. Being able to bypass this histology phase of diagnosis and to get answers directly from thick specimens within minutes would be welcomed by both clinicians and patients.
In a recent advance, researchers at the University of California, Davis, along with Stavros Demos, Ph.D., now at the University of Rochester, have developed a new technique that speeds up the diagnostic process by skipping most or all of these traditional histology steps. Microscopy with UV surface excitation (MUSE) takes advantage of the fact that the penetration depth of deep UV light (around 280 nm) is limited to approximately 10 µm, just slightly thicker than conventional histology slides.
This effectively restricts the excited region of the briefly stained samples to the surface and provides an optical alternative to physical sectioning. This is in contrast to visible-wavelength excitation, which has greater depth penetration and thus results in considerable out-of-focus fluorescence signals. MUSE images from cervical and skin epithelium as well as connective tissue show the clarity of the technique (Figures 1 and 2).
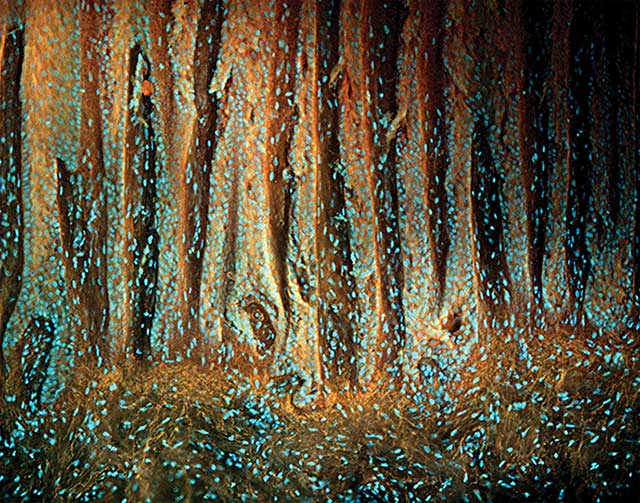
Figure 2. Abundant growth of skin epithelium in a plantar wart. Orange elastin fibers are visible below the epithelial layer. Courtesy of Richard Levenson.
There are currently other techniques that seek to avoid the need for prior tissue sectioning by directly examining the surface of fresh specimens. The common theme in all of these approaches is to reject the light from unwanted layers of the tissue during the imaging. Confocal microscopes use a pinhole. Multiphoton scanning microscopes rely on the nonlinearity of a two- (or more) photon excitation process that limits the illuminated area to a tightly focused region. Optical coherence tomography uses interference of reflected light with a reference beam to select signals from a desired depth. More recently, structured illumination and light-sheet microscopy also have demonstrated utility in this area.
MUSE is optically simpler than these other approaches. It is a variant of wide-field (nonpoint-scanning) microscopy that uses an illumination wavelength of 280 nm to excite tissues stained briefly (seconds) with a number of common, inexpensive fluorescent dyes. The LED light source is positioned adjacent to the objective to obliquely illuminate the sample (Figure 3). Standard collinear (i.e., through the lens) illumination using a typical epifluorescence optical path is not useful for MUSE, as typical lenses are opaque in this UV wavelength range.
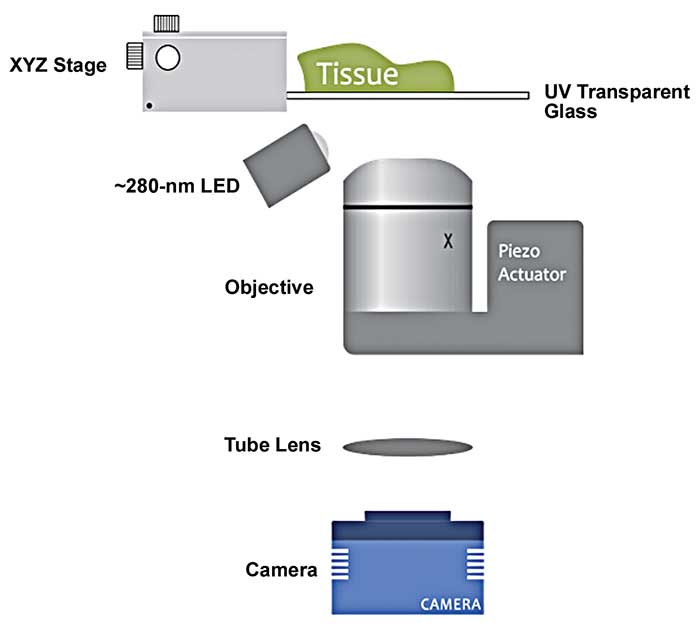
Figure 3. A diagram displaying the optical setup for a MUSE (microscopy with UV surface excitation) microscope. The tissue is supported on a UV-transparent window connected to a XYZ-stage. A 280-nm LED excites the specimen obliquely, generating visible-range emission that is directed by a conventional microscope lens to a color camera. Courtesy of Richard Levenson.
The arrival of the excitation light at the sample from outside the objective does have benefits. It removes the requirement for a dichroic mirror in the light path and, as the lens itself blocks UV light, there is no need to introduce an additional excitation filter. However, accommodating an LED source close to the sample can interfere with properly positioning short-working-distance objectives, which currently limits the choice of lenses to those with relatively long working distances.
Many fluorophores that emit in the visible spectral range share a common (but not widely appreciated) excitation regime at wavelengths below 300 nm, associated with high-energy excitation from ground state (S0) to the second excited state (S2). This is in contrast to the more familiar S0-S1 excitation, where the fluorescent molecules emit photons at wavelengths not far from those used for excitation (i.e., they exhibit small Stokes shifts).
By using a single, 280-nm UV excitation source, MUSE can simultaneously excite multiple fluorescent reporters in the sample to achieve multicolor fluorescence imaging without the hardware complexities associated with most conventional multichannel fluorescence microscopes. For example, eosin — a common histology dye that has useful fluorescence emissions — has an S1 absorption peak around 520 nm. Excitation light in this range results in green fluorescence peaking at approximately 540 nm (Stokes shift of ~20 nm). However, when excited at 280 nm, it emits with the same spectrum, peaking at 540 nm, thus exhibiting a Stokes shift of 260 nm.
Since a number of common dyes share this property, it is possible to employ as many as four or more stains simultaneously. As narrowband emission filters are not required, the signals can be collected with a simple color camera to generate multihued and informative images. A spinal cord, for example, can be stained with two conventional fluorescent dyes, rhodamine B and Hoechst (Figure 4). While these dyes have usual excitation wavelengths of about 550 nm and 350 nm, respectively, they both fluoresce with considerable brightness when excited at 280 nm.
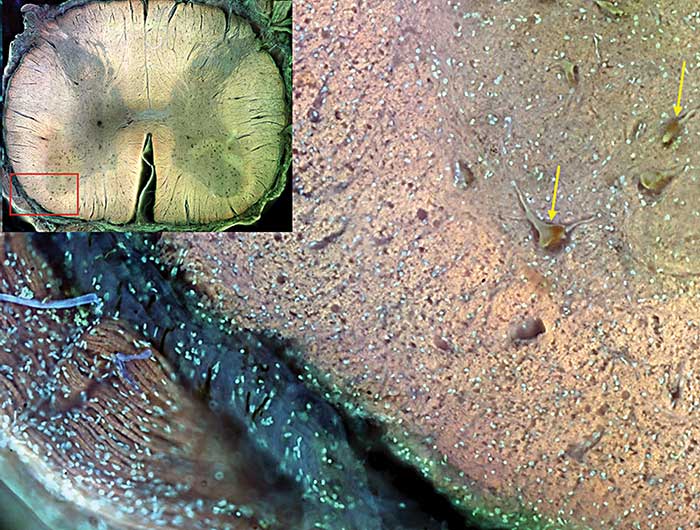
Figure 4. Inset is a stitched montage of a cross section of an adult human spinal cord imaged with MUSE. In the bottom left corner of the main image, peripheral nerve is seen. Leptomeninges characterized by autofluorescent blue collagen are also visible. Yellow arrows point to motor neurons in the spinal cord itself. Courtesy of Richard Levenson.
MUSE owes its practicality to the relatively recent availability of LED-based UV light sources with the necessary efficiency, available power and deep-UV range. As LED sources continue to become cheaper and brighter, the requirement of high-NA lenses and the restriction of objective choice based on working distance will decrease. A 280-nm LED is the preferred option for cost and performance reasons, but as additional sub-280 nm LEDs come to market, there may be more optimal choices.
Eliminating the need to fix, embed and physically thin-section patient samples can greatly reduce time to diagnosis, as well as the use of specialized histology equipment and toxic solvents. However, speed is not the only metric; it is also important to replicate the appearance of conventional hematoxylin and eosin (H&E) staining and thereby increase acceptance from microscopists, including pathologists, who are used to interpreting the pink and blue colors of H&E. Fortunately, it is possible to convert from native MUSE mode to simulated H&E in real time (Figure 5). The ability to display the two modes simultaneously can help viewers adjust to and interpret both the original fluorescence and the simulated bright-field display, as certain features are better shown in one or the other.
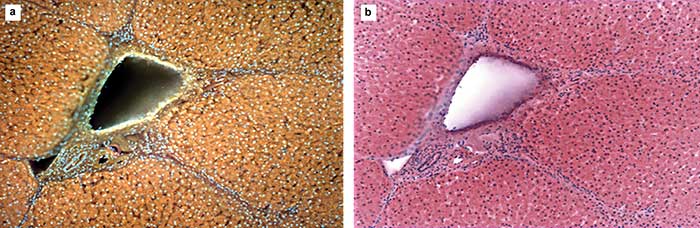
Figure 5. Panel a shows fixed porcine liver imaged with MUSE. Pictured is a large portal vein with associated bile duct and arteriole. Panel b shows a virtual S0-S1 excitation, generated from the image in panel a. In the native fluorescence image, the depth of the holes is evident, while the depth is lost once the color inversion is complete. Courtesy of Richard Levenson.
With the advent of precision medicine efforts, the morphology of the tissue specimen is now just the beginning of the process. However, diagnosticians are confronted with increasing numbers of complex molecular diagnostic tests that must be accommodated by ever-smaller tissue samples. Such small samples may prove to be the only specimens on which conventional microscopy can be performed, via either frozen section or the more usual formalin-fixed paraffin-embedding (FFPE)-based processes, either of which can consume most or all of the scarce sample, leaving little or nothing left over for molecular testing.
Moreover, FFPE and exposure to different solvents can all adversely affect the integrity of protein and nucleic acid analytes. As MUSE is not only nondestructive, but also avoids these potentially deleterious steps, it could be an excellent preamble to molecular testing. Preliminary studies have demonstrated no adverse results of prior MUSE imaging on subsequent RNA-seq, FISH (fluorescence in situ hybridization) and immunohistochemistry (IHC) assays.
While MUSE has a lot to offer in terms of speeding up the diagnostic process, or in fact for allowing for diagnosis to happen at all in low-resource settings, it also provides images not achievable using conventional histology. Because there is no sectioning, and no loss of thickness, the technology often yields more informative scenes. The process, for example, can reveal the complex organization of collagen fibers in the canine dermis (Figure 6). Much of this visual content would be lost if this specimen had been conventionally sectioned.
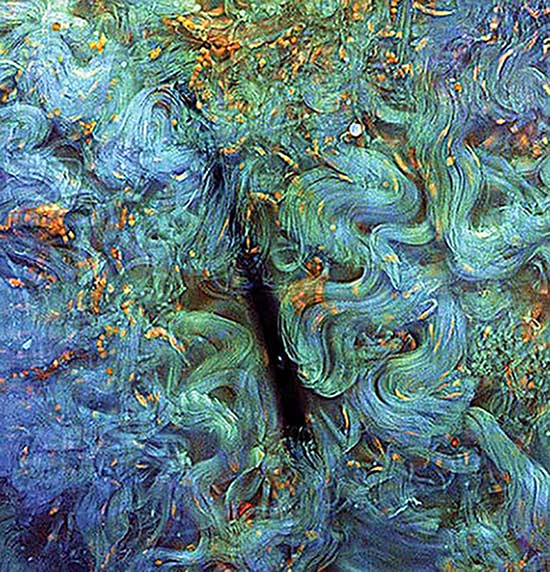
Figure 6. Malignant canine mast cell tumor. Orange mast cells are embedded in a dense and complex arrangement of collagen fibers. Courtesy of Croix Griffin, UC Davis.
This research was published recently in an article titled “Microscopy with ultraviolet surface excitation for rapid slide-free histology” in Nature Biomedical Engineering (doi:10.1038/s41551-017-0165-y).
Meet the authors
Farzad Fereidouni is a project scientist in the Department of Pathology and Laboratory Medicine at UC Davis Health. He obtained his Ph.D. from Utrecht University in the Netherlands and is an experimental physicist, experienced with imaging and microscopy; email: [email protected]. Austin Todd is a research volunteer in the Department of Pathology and Laboratory Medicine at UC Davis Health, having recently graduated in cell and molecular biology from California State University, Sacramento; email: awtodd@
ucdavis.edu. Richard Levenson, M.D., is professor and vice chair for Strategic Technologies in the Department of Pathology and Laboratory Medicine at the University of California, Davis. He previously served as VP for Research at Cambridge Research and Instrumentation (now part of PerkinElmer) and works at the intersection of optics, software and medicine; email: [email protected].