From watching embryos as they develop to viewing neurons being manipulated in the brain, live-cell imaging continues to find new applications.
Since its introduction in the 1600s, improvements in microscope technology have continually broadened the types of cells and cellular processes that can be studied. Advances in automation have made this already-simple tool faster and more capable, and time-lapse imaging reveals function and dynamics in addition to structure.
Live-cell imaging has enabled us to witness incredible moments in biology in unprecedented detail. Even embryogenesis – the process of cell division and cellular differentiation that occurs at the earliest stages of life – has recently been captured.
Much of what we know about the cell cycle, chromosome segregation errors and the development of cancer has come from live studies of cells exposed to small molecule inhibitors and other drugs.
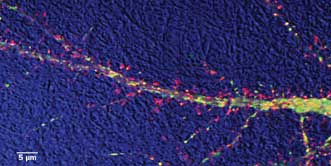
Study of dendritic transport in hippocampal neuron: DIC (bright field) detail is in blue; actin protein
is labeled with red; and an Alzheimer’s disease-related protein is in green. Courtesy of Virginie
Buggia-Prevot and Celia G. Fernandez, Gopal Thinakaran lab.
In fact, questions of why diseases take hold and progress are a major driving force pushing scientists to improve live-cell imaging techniques. As well as many types of cancer, scientists are keen to unravel the inner workings of neurodegenerative diseases such as Parkinson’s or Alzheimer’s.
Live-cell imaging can be divided into bright-field and fluorescence image capture. Apart from documenting changes in cell shape or cell motion, both technologies allow for specific and diverse types of function measurements.
“In bright field, the sample is trans-illuminated, and the density of the cell creates contrast that is used to observe features and dynamics,” said Dr. Vytas Bindokas, facility and technical director of the Microscopy Core Facility at the University of Chicago. “Bright-field methods are among the oldest techniques, but still highly useful.”
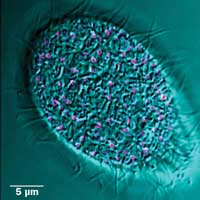
Fluorescence image of mucocyst tip protein (pink) and DIC bright field (cyan) in Tetrahymena. Moving cells pose a challenge for imaging: Simple cells have long served as models for study of similar processes in more complex organisms. Preparation courtesy of Santosh Kumar, Aaron Turkewitz lab.
Fluorescence methods are extremely versatile and can track multiple specific types of information at once. By applying one or more of the ever-growing palette of fluorescent proteins now available, a bright signal is created against a dark background.
“Vital fluorescent dyes in numerous colors are available to label cellular compartments and to read out physiologic processes in real time,” Bindokas said. “Examples of physiologic measurements that are associated with major cell events include changes in pH, ion concentration, membrane voltage, enzyme activity, generation of reactive molecules/species, etc.”

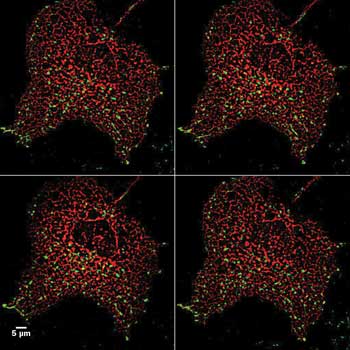
Vital staining of mitochondria (red) within fibroblast cells (gray). Dynamics of these fragile, energy-producing organelles are recognized as being increasingly important in disease. Preparation by Sadhana Samant, Mahesh Gupta lab.
Superresolution for live-cell imaging
One of the biggest trends at the moment is the tremendous effort under way to get superresolution techniques working under live-cell conditions.
“For many years, it was not possible to perform any ‘real’ live-cell experiments under quasiphysiological conditions,” said Sebastian Tille, director of wide-field imaging at Leica Microsystems GmbH in Germany. “This has changed and delivered new insights. For instance, it was possible to follow the movement of dendritic spines with STED [stimulated emission depletion]. Also, 3-D superresolution (localization) microscopy allows going far beyond the diffraction limit in all three spatial dimensions down to the molecular level – now possible with the Leica SR GSD 3D.”
3-D projections of mitochondria in cancer cells: Recent studies suggest mitochondria may be targets for controlling cancer. Preparation by Rifat Hasina, Ravi Salgia lab.
Images are now being produced with detail previously only attained with the electron microscope. With superresolution, the limit for visualization now approaches the size of common proteins.
Structured illumination microscopy (SIM) and stochastic optical reconstruction microscopy (STORM) are two superresolution techniques that have advanced significantly in recent years, with some impressive results obtained from living cells.
SIM uses wide-field illumination of the specimen with patterned excitation to create interference patterns called moiré fringes. These fringes contain additional information that can be used to generate a superresolution image with a twofold improvement in resolution over conventional fluorescence techniques.
Time-lapse imaging of green fluorescent protein-prion protein in cells expressing the red fluorescent protein DsRed within the endoplasmic reticulum. Prions can cause neurological diseases that include Kuru and mad cow disease. Preparation from James Mastrianni lab.
“We are seeing some amazing live-cell SIM images coming from the labs of Hari Shroff at the NIH [US National Institutes
of Health] and Eric Betzig of the Howard Hughes Medical Institute,” said Dr. Christopher O’Connell, manager of superresolution system products at Nikon Instruments Inc. in Melville, N.Y. “These researchers are using novel methods of generating structured illumination to advance SIM technology to the next level, creating systems with incredible temporal resolution and the ability to image at depth. Even whole embryos can be rapidly scanned with these methods.
“STORM is a form of localization microscopy that cumulatively maps the fluorescence from individual dye molecules to create an image with ten times better resolution. Both SIM and STORM are allowing researchers to observe structural features that were previously only visible by electron microscopy.”
Other techniques
While high-speed confocal imaging of fluorescent reporters seems to be the most common and powerful tool being used today for live-cell imaging, there are other specialized techniques that are used to study cellular membranes and events.
Total internal reflection microscopy, for example, uses a high-numerical-aperture objective lens and a spot of laser light to make the laser reflect off the glass-cell interface and back into the lens. Since the technique involves shallow illumination, photo damage to the sample is limited.
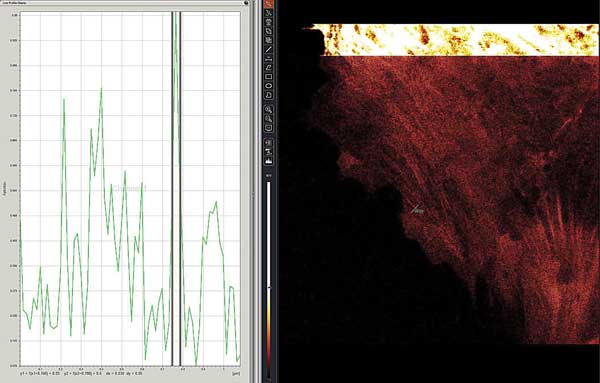
Live-cell STED-CW superresolution of green fluorescent protein-MLCK protein in lung endothelial cells. Region intensity plot shows 39-nm FWHM (full-width half-maximum) diameter for a stress fiber indicated as ‘ROI1’ and green line segment near center image. The bright, upper inset shows the same cell in standard confocal mode. Preparation courtesy of Mary Brown, Steve Dudek lab.
A product that has only just reached the market for whole- or small-organism imaging is light-sheet microscopy. This is similar to a point-scanning confocal system, except that the scanning is carried out by a thin sheet of light; image capture is typically at right angles to the plane of illumination. This is another approach that causes very little photo damage because it uses high-speed cameras and low light, but the labeled cells of the entire organism can be tracked over time.
Another technique known as spinning-disc microscopy continues to hold a lot of power for those looking at monolayer cells because of the high imaging speed and resolution that are possible when using the latest generation of cameras available on the market.
In this approach, hundreds of spaced excitation pinholes rotate at high speed to sweep light across the sample so as not to interfere with each other’s signals. The image can then be captured using high-sensitivity digital cameras that include some of the best-efficiency photodetectors available today (up to 93 percent efficiency).
“We recently introduced a new Flash4 sCMOS camera for the UltraVIEW VoX 3-D live-cell imaging system that improves sensitivity and increases frame rates,” said Dr. Jacob Tesdorpf, director of high-content instruments and applications at PerkinElmer in Hamburg, Germany. “This enables scientists to capture more images per second, gaining sensitivity and speed, and resolving intracellular process faster than before.”
The challenge of imaging the living
Living cells pose a unique challenge when it comes to microscopy. Firstly, limiting light exposure continues to be one of the main challenges. Of course, light is crucial to imaging, but too much light can manifest in subtle ways to influence research results; it can even be damaging to the cell.
“Fluorescence illumination, especially in the UV range, is harmful for cells and causes photobleaching and phototoxicity,” Tesdorpf said. “It is therefore important to make every photon count, by limiting unnecessary excitation to a minimum and capturing a maximum of emitted photons.”
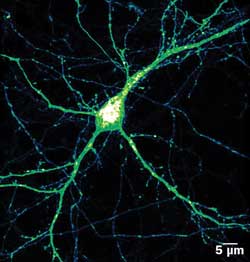
Expression pattern (intensity color coded) of BACE1-yellow fluorescent protein in mature hippocampal neuron. BACE1 is involved in Alzheimer’s disease. Its movement can be tracked from the bulbous cell’s body, through the thicker dendrites and even the finest axons. Courtesy of Virginie Buggia-Prevot, Gopal Thinakaran lab.
The problem is particularly true for superresolution methods, which require more raw information than conventional fluorescence methods. This additional information comes from extra images or high laser power needed to selectively turn fluorophores on and off.
Apart from light toxicity, mammalian cells need to be kept at 37 °C and may require elevated levels of carbon dioxide to maintain vigor.
Various incubation schemes exist as add-ons to standard microscopes that are reported to work with varying levels of success.
Imaging normal cell dynamics within whole animals, especially the mouse model, is also challenging due to the depths required for image capture. Whole-animal imaging requires repeat imaging at the same location while preventing infection and inflammation, and accounting for movement arising from the heartbeat and respiration of the animal.
On the horizon
Recently, a few commercial imaging microscopes have been built inside standard cell incubators to permit live imaging in an ideal cell environment. Some include provisions for multiple chambers of cells to allow concurrent experiments to be run as well as the ability to introduce drugs/agents in an automated fashion.
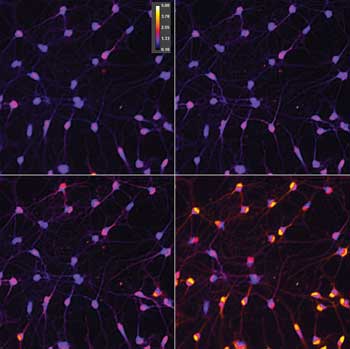
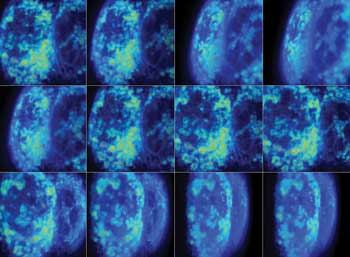
Measurement of intracellular free-calcium content in neurons using fluorescent ratiometric reporter. The color-coded ratio value shows a calcium rise as hotter color values. These images show that responses are not equal in all cells, with some responding faster than others. Preparation from Jeremy Marks lab.
Major brands of microscopes now also offer an option to maintain the focal position of an in vitro sample by means of an optical feedback. This allows long-term imaging even when solutions are changed in the sample dish.
Supercontinuum (white light) lasers have recently been made available for confocal microscopes. This means excitation probes can be freely selected from a complete spectrum of excitation lines, and they can be finely tuned as needed.
Finally, time gating is now being applied to improve superresolution techniques. Gated-STED offers further resolution gains with potentially lower light requirements.
“Use of a delay between the excitation pulse and the onset of emission collection time window offers rejection of light contamination from the excitation laser and the portion of fluorescence that initially failed to be depleted by the depletion laser, thus improving the resolution,” Bindokas said. “Live-cell imaging is becoming more turnkey, but it’s not there yet. Each preparation requires optimization to extract accurate data versus potential artifacts.
“While this aspect is no different than for any scientific protocol, a successful live-cell imaging has one advantage over many techniques: It can result in beautiful images plus data.”