The microscopy technique uses multiple, sequentially fired lasers in conjunction with software algorithms to capture and map cellular details.
Scientists and clinicians have long believed that visualizing structural changes in the nucleus of cancer cells would unlock vital clues about how the disease first forms. This information could provide the ability to track where and how cancer is progressing to plan its successful treatment. Traditional microscopy methods have fallen short in revealing these details because the microscope’s inherent resolution limit only allows the imaging of general structural changes, as opposed to resolving their point of origin. Finding fluorescent dyes that illuminate abnormalities, such as aggregated proteins, at the single-molecule level and effectively target their spatial distribution has also not been easy. Innovations in superresolution technologies such as stochastic optical reconstruction microscopy (STORM) are helping life science researchers to understand how healthy and diseased cells function at their core.
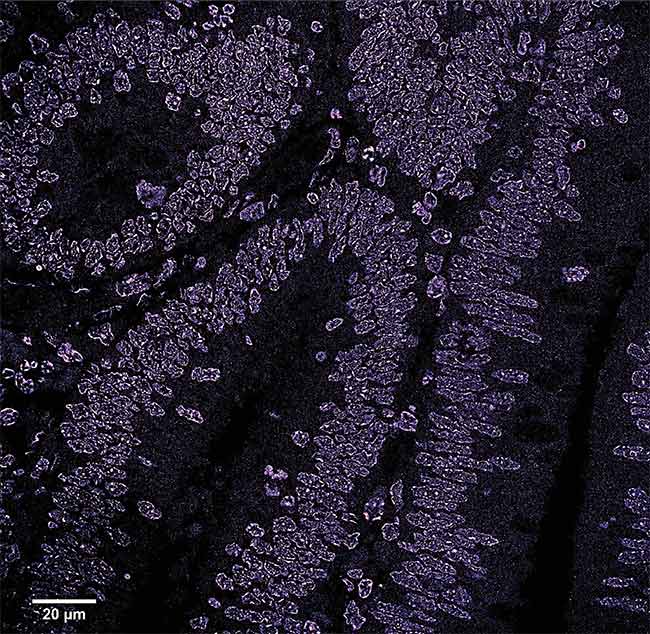
A stochastic optical reconstruction microscopy (STORM) image of a region of a colon polyp. The image shows the disruption of the chromatin structure in the early stages of cancer. Courtesy of the University of Pittsburgh.
STORM is a single-molecule localization technique in which a subset of labeled fluorescent molecules are activated by laser light while others remain dark. This process is repeated until a full picture is created that differentiates fluorescent molecules with nanometer accuracy. Yang Liu, an associate professor in the Department of Bioengineering at the University of Pittsburgh, and her colleagues used the technique in their research to examine the chromatin — the materials that make up chromosomes, including RNA, DNA, and proteins — of cancer cells (Figure 1).
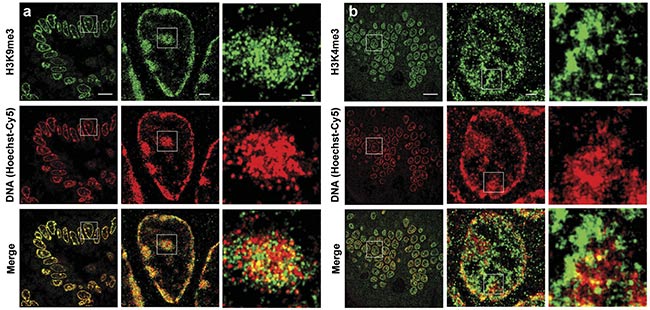
Figure 1. Two-color superresolution images of DNA and histone marks on mouse intestinal tissue. STORM images show the spatial relationship between DNA and the heterochromatin regions marked by H3K9me3 (histone 3 lysine 9 trimethylation) (a). STORM images show the spatial relationship between DNA and euchromatin regions marked by H3K4me3 (b). Scale bars: 10 µm, 1 µm, and 200 nm, respectively, in both the original and magnified images. Courtesy of the University of Pittsburgh.
“We focused on the mostly densely packed DNA or heterochromatin in the cell nuclei. Heterochromatin is considered to be the guardian of the genome, as it is essential for maintaining genomic stability,” Liu said.
Heterochromatin is a condensed form of chromatin that is packaged in a hierarchical way within cells, and it regulates accessibility to DNA, which could be affected by various processes during an organism’s life cycle (Figure 2).
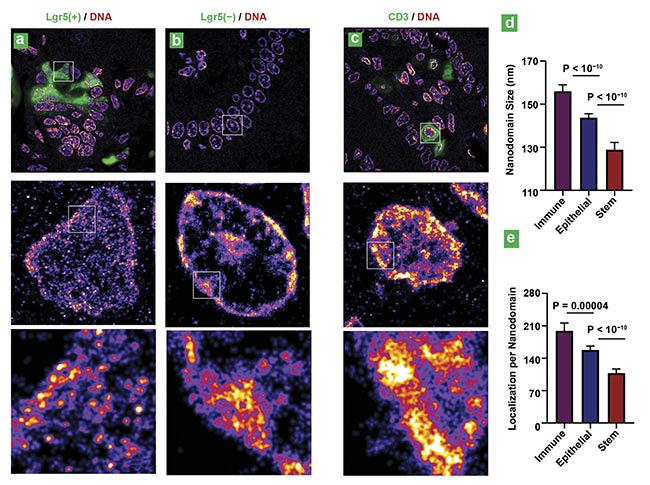
Figure 2. Superresolution imaging of genomic DNA in various cell types from mouse intestinal tissue. Superresolution images of DNA from intestinal stem cells (Lgr5 positive) and differentiated cells in the villi region (Lgr5 negative) (a, b). Superresolution images of DNA from immune cells marked by CD3 (cluster of
differentiation 3) (c). Statistical analysis of the DNA nanodomain size and the number of localizations per nanodomain for each group (d, e). P: probability.
Courtesy of the University of Pittsburgh.
“Abundant evidence shows that disrupted heterochromatin leads to genomic instability — an ‘enabling characteristic’ for cells to acquire hallmarks of cancer,” Liu said. “We found that visualizing disrupted heterochromatin structures at nanoscale resolution [could enable the ability to] detect the early-stage malignant transformation even before the tumor is formed.”
In a published study, Liu and her colleagues in her lab used a small-molecule fluorescent probe — cyanine 5-conjugated Hoechst, or Hoechst-Cy5 — in conjunction with an Olympus IX71 inverted microscope frame and a 642-nm excitation laser to create STORM images of tissue, particularly of the folding of the chromatin structure, at various stages of cancer progression with nanoscale resolution. The researchers observed a variety of changes in the chromatin of cancer cells, such as the disruption of chromatin compaction in the early stages of colon cancer. This disruption results in the breakdown of the folding over time. The level of breakdown varied between the chromatin in healthy and cancerous colon cells. The chromatin was fully compacted in the healthy cells, while in the cancer cells it was highly disrupted and assumed an open structure1.
The team also discovered that this disruption was universal among many types of tumors — including colorectal, pancreatic, prostate, lung, and ovarian cancers — which could ultimately identify key characteristics of cancer’s genesis that are not captured by traditional pathology.
When Liu and her team started their work in 2014, they used a commercially available STORM system called N-STORM, which at the time took about 20 min to image one nucleus and several hours to reconstruct a full image of tissue. This duration was not suitable for full-scale clinical analysis, she said. More modern versions of the system, however, can image hundreds of nuclei in about 10 min.
“STORM was initially developed as a basic research tool to visualize molecular-scale structure in biological samples and now has become a standard tool in cell biology and other biomedical research,” Liu said. “From the standpoint of the hardware, STORM has a great potential to be used as a clinical tool due to its nanoscale resolution down to about 20 nm, the simplicity of the instrumentation, and its relatively low cost compared to other high-end microscopy systems.”
To make the STORM method applicable for diagnostics, structural features such as chromatin folding must be superresolved in a robust manner to collect and process samples from patients in the standard clinical setting. In the future, she said, this could help to identify patients at high risk for cancer or to determine which lesions are benign and which are malignant.
Within the context of a wave of superresolution methods that have broken the historical optical diffraction limit, one of the initial descriptions of STORM was provided by a group at Harvard University that included Xiaowei Zhuang, Michael Rust, and Mark Bates. They described the STORM method as a series of imaging cycles, during which only a fraction of the fluorophores are activated by laser excitation in each cycle. This process produces an image with a 20-nm (or higher) lateral resolution2.
The utility of this technique was shown in the laboratory at around the same time that Eric Betzig and his colleagues at the Howard Hughes Medical Institute were establishing similar methods such as photoactivated localization microscopy (PALM) (see sidebar).
Gathering STORM
STORM experiments are often accomplished by using an inverted microscope, although results can be obtained with other designs, such as a light-sheet microscope. With an inverted epifluorescence microscope, the objective lens is located below the sample and points upward, projecting excitation light and collecting fluorescence signal through the bottom of the specimen chamber.
To suppress the background fluorescence, the excitation light is often set at a high incident angle close to but smaller than the critical angle, said Bo Huang, a professor in the Department of Pharmaceutical Chemistry and Biophysics at the University of California, San Francisco.
“STORM and PALM are very similar in that they are both based on collecting signals from single molecules,” Huang said. “But what is created is not based on pixels but point coordinates that are then rendered into an image.”
At the Abbe limit, proteins can appear blurry because many of them may be clustered together. However, with STORM, the overlapping of point spread functions can be avoided and the proteins’ coordinates can be marked by fitting a Gaussian function to the signals that are collected.
From the earliest innovations that launched this technique, variations of STORM, depending on the parameters of each experimental setup, have been introduced.
One particularly common variation is called direct stochastic optical reconstruction microscopy (dSTORM). This technique does not require the use of a tandem fluorescent dye but instead uses more powerful (2- to 4-W) pulsed diode lasers that excite commonly available “blinking” fluorophores.
The practicality of dSTORM
“dSTORM is easier to do both on the sample prep side and acquisition/setup side and has no performance/resolution disadvantages over classical STORM,” said Rob Davison, market development specialist at Thermo Fisher Scientific. “There may still be unique applications out there where lower laser power is necessary, but live-cell/tissue imaging isn’t one of them, as the typical STORM acquisition requires hundreds to even thousands of sequential frames of blinking fluorophores to create an image. Living, dynamic samples don’t lend themselves well to this. But there is a trade-off: As more frames means better localization data and a better resulting image, it also takes more time.”
The instrumentation that is needed to take advantage of this technique has fortunately become increasingly accessible to researchers and scientists.
“You can buy 500-mW to 1-W lasers for $5,000 to $10,000 now, when they cost well over $30,000 10 to 15 years ago,” Davison said. “This has led to almost everyone going to dSTORM over traditional tandem-dye-based STORM over the past five or 10 years.”
Nevertheless, he said, much of the technology inherent to dSTORM has changed little in recent years, other than small adjustments that have led to improvements in the field of view (FOV). And, in general, STORM techniques work best with very thin samples, such as cultured or plated cells. Tissue exhibits substantial autofluorescence and is thick, which is not advantageous when using high laser power.
“Optics on STORM systems typically don’t allow much more than 200 nm of imaging depth. And though there are some tricky optical physics [that can be used] to get beyond that depth, they aren’t widely used yet and they require even longer acquisition times,” Davison said. “Sample prep is also more involved than STED/SIM [stimulated emission depletion or structured illumination microscopy], which presents another challenge, on the clinical level, for creating a validated screen/assay.”
Speedy image acquisition
No matter which iteration of superresolution microscopy is used, the inherent advantages of using such high resolution include the ability to probe structures as they change. By resolving individual proteins at nanoscale resolution and counting them, as well as analyzing their spatial distribution and movement, it becomes possible to understand the cell morphology at a level of detail largely unseen in traditional optical microscopy methods.
Biotech company ONI offers a microscopic imaging system called the Nanoimager (Figure 3) that is capable of resolution greater than 20 nm (Figure 4). The technology enables visualizing, tracking, and imaging single molecules in live and fixed samples. Up to four lasers — which excite specific fluorophore colors at 405, 488, 560, and 640 nm — can be utilized by the technology. Both full-frame and compressed modes are available, the latter of which can substantially improve imaging speed.

Figure 3. The Nanoimager microscopic imaging system can be used for a variety of superresolution techniques. Courtesy of ONI.
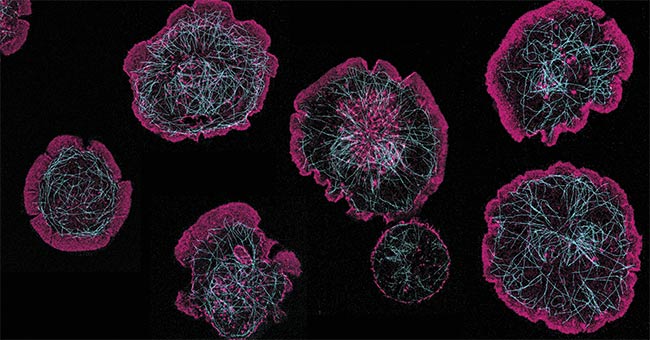
Figure 4. Rat basophils activating on a surface coated with rat immunoglobulin E. When the cell activates, actin (stained with Phalloidin-Atto 488) (pink) flows to the edges and causes the cell to spread and form a large contact with the glass surface. The microtubule network (stained with anti-tubulin Alexa Fluor 647) (cyan) reorganizes to facilitate the release of proinflammatory factors (e.g., histamine), which occurs during an allergic response. Courtesy of James Felce/ONI.
One of the many applications of superresolution is the characterization of extracellular vesicles, which are a group of membranous structures that includes exosomes and microvesicles. Extracellular vesicles exist in biological fluids and are integral to a variety of physiological processes. They are also linked to interactions between cells, such as the exchange of proteins, lipids, and genetic material.
The more that is learned about the origin and ultimate fate of these vesicles in the body, the more that research scientists believe they will be able to understand about the basics of human and animal biology. This understanding could open the door to knowledge about the genesis of cell functions and — most significantly, when it comes to potential clinical application — how these cellular processes can break down, along with providing diagnostic tools to track when patients are at risk.
“This type of structural analysis, below the diffraction limit, can help capture signaling receptors in neuroscience, or the mechanisms behind metastasis in cancer research,” said Jason Jell, ONI’s vice president of marketing. “This [analysis] has been mostly used in the life sciences, but it has real implications for diagnostics. If you look at most techniques for performing diagnostics, what is being considered is an average of data found at a particular place and time. But there is a lot of heterogeneity in people, and superresolution can help establish the peculiarities in each individual.”
He said one major advancement in recent years in the field of superresolution microscopy has been the explosion of software that is available to researchers both inside and outside the laboratory. While ONI offers its own software to accompany its Nanoimager, other packages are on the market.
“The picture that STORM makes possible is really built with an algorithm,” Jell said. “But what this software enables is the separation of a researcher or clinician from the equipment, which means that a sample can be prepared with the actual analysis occurring outside the laboratory.”
This capability could allow for a broader understanding of drug delivery in pharmaceutical tests, he said. “Yes, superresolution and STORM are principally at use in life sciences research. But, in my view, we are just one assay away from establishing this technology in a clinical setting. It offers a whole different level of sensitivity.”
A changing view
Another company that works in the arena of superresolution enhancements to fluorescence microscopy is Bruker. The company works with single-molecule modalities such as STORM, in particular. After acquiring Vutara (a company specializing in 3D superresolution microscopy) several years ago, Bruker has recently focused on collaborating with researchers in the study of DNA structure, messenger RNA (mRNA) distribution, and subcellular compartments.
To strengthen this effort, Bruker incorporated a variety of tools into the Vutara VXL superresolution workstation, which encompasses a compact imaging system that achieves about 20-nm resolution. The system uses a patented technology called “biplane,” which essentially means imaging two focal planes at once in relation to a sample. This makes it possible to determine the axial position of a particle.
This principle was tested with an inverted microscope setup and five high-powered solid-state lasers, along with a silicon oil immersion objective. With the aid of mirrors and a beamsplitter cube, light can be projected to two distinct locations relative to the sample3.
The device includes a sCMOS camera and five colors that can be acquired via the five excitation lasers at wavelengths between 405 and 750 nm. It can be combined with a microfluidics unit to enable experimentation in spatial genomics, transcriptomics, and proteomics. The technology also holds promise in virology and neuroscience, Bruker product manager Winfried Wiegraebe said.
“Normally, in microscopy, you put a camera directly in the focal plane, but with this setup you split the image into two FOVs so that it is slightly out of focus. With this setup, we can acquire 3D images in thick samples not accessible with other technologies,” he said. “Most people are limited in the number of structures they can image by the number of probes they can spectrally differentiate. We overcome this limitation with microfluidics, multiplexing, and barcoding technology like DNA-PAINT.”
In the DNA-PAINT (point accumulation in nanoscale topology) technique, the transient binding of short dye-labeled DNA or RNA strands to their target results in a “blinking” effect that helps to accomplish the STORM technique.
According to Wiegraebe, high laser power is vital in techniques such as dSTORM to drive certain fluorescent molecules into a dark state, since lower-power lasers would necessitate longer exposure times. The higher power allows for overcoming increased artifacts or photobleaching.
From lab to clinic
Up until now, different varieties of STORM have proved to be valuable to basic biological research in studying chromatin structure and the relationships between various cellular organelles, with companies and academic institutions focusing on life science research. While the technique holds promise for clinical applications when the building blocks of various diseases are identified, Wiegraebe said some practical hurdles remain.
“Right now, the throughput is too slow for most diagnostics, which requires results as close to real time as possible,” he said. “These systems are in the early stages of research and development in the discovery of effective drugs in disease treatment. When you’re studying cellular structures, a large amount of data is collected which needs to be stored and analyzed.”
The future of superresolution techniques will likely be governed by a number of disciplines working together toward developing both data analysis software and instrumentation to make the technology available in as many settings as possible.
Burgeoning trends include using adaptive optics to correct aberrations, which are diversions of the light path caused by the components of the microscope (such as lenses or mirrors) or by various materials within the sample itself.
“In my view, every microscope could be designed with adaptive optics, because the aberration can be corrected without changing the whole microscope setup,” said UC San Francisco’s Huang.
Photoswitchable fluorescent dyes that enhance specificity and photon output have been created to improve the resulting image. Scientists are also working to use the various branches of superresolution in the same experiments, such as in the sequential use of structured illumination microscopy (SIM) and single-molecule localization microscopy (SMLM), to provide both structural and single-molecule information.
Improvements in AI will build a link to the data obtained through parallel research conducted using other microscopy technologies. This expanded knowledge will inform both research and diagnostics in fields such as neuroimaging and could enable the characterization of cancer cells and other diseased cells.
“Looking forward, what you will see is fewer publications about the design of superresolution imaging systems and more publications about the collection of data and what it tells us about biological systems,” Huang said.
References
1. J. Xu et al. (2022). Ultrastructural visualization of chromatin in cancer pathogenesis using a simple small-molecule fluorescent probe. Sci Adv, Vol. 8, No. 9, p. eabm8293.
2. M. Rust et al. (2006). Stochastic optical reconstruction microscopy (STORM) provides sub-diffraction-limit image resolution. Nat Methods, Vol. 3, No. 10, pp. 793-795.
3. M. Mlodzianoski et al. (2009). Experimental characterization of 3D localization techniques for particle-tracking and super-resolution microscopy. Opt Express, Vol. 17, No. 10, pp. 8264-8277.
Breaking the Abbe Limit: Two Decades of Superresolution
Superresolution microscopy became possible in practice with the breaking of the so-called Abbe limit. This limit involved the fundamental principle — established by microscopist Ernst Abbe in 1873 — that the resolution of an optical microscope could never exceed 0.2 µm, or half a wavelength of light. Beyond this threshold, objects would bleed together in the view under a microscope’s lens.
Around the year 2000, a variety of techniques were developed to overcome this limitation, with the aid of fluorescent dyes to capture specific molecular details, separated by space and time, in a sample.
These advancements ultimately led a trio of scientists — Eric Betzig of the Howard Hughes Medical Institute (HHMI), Stefan Hell of the Max Planck Institute for Biophysical Chemistry in Gottingen, and William Moerner of Stanford University — to receive the Nobel Prize in chemistry in 2014. Hell is credited with developing stimulated emission depletion (STED) microscopy, in which two laser beams are used, one to excite all of the fluorescent molecules in a sample and the other to remove the fluorescence except from a tiny area. Meanwhile, Betzig and Moerner are acknowledged for conceiving of single-molecule microscopy, which uses a laser to activate (and deactivate) the fluorescence of specific molecules.
Betzig and his colleague Harald Hess at HHMI are credited, in particular, with inventing photoactivated localization microscopy (PALM), which identifies individual molecules through stochastic excitation via a lower-power laser beam and reconstructs an image over hundreds of frames.
It was in this context that stochastic optical reconstruction microscopy (STORM) was conceived. The STORM method utilizes a series of imaging cycles, and only a fraction of the fluorophores are activated in each cycle. The cycles are repeated until a full picture is created that differentiates fluorescent molecules with nanometer accuracy. This process produces an image with a 20-nm (or higher) lateral resolution.
Also added to the microscopists’ toolbox was a method called structured illumination microscopy (SIM), which involves the excitation of patterned illumination — to be followed by computational unmixing — to construct fluorescence images in 2D or 3D. Subsequent changes to all of these methods generally revolved around the needs of researchers in biology, whether in spatial or temporal resolution.