Total internal reflection is the curious interaction of light and matter that makes fiber optic telecommunications possible, but its use in biological imaging may be even more interesting still.
In any method of fluorescence microscopy, control over which particles fluoresce and which do not is a key challenge. For the most part, the struggle is to get the minimum amount of fluorophores to emit light – at just the right time – before snapping your subject’s photo. Too many particles glowing in the background, beyond the focal plane, create unwanted noise, obscuring the image and accelerating, in some cases, possible photobleaching effects.
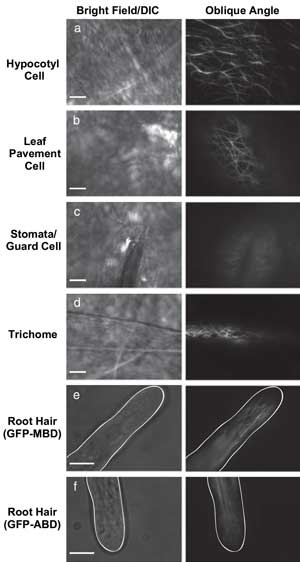
VAEM is useful for imaging fluorescent protein located in a variety of plant cell types. Courtesy of The Plant Journal.
In microscopy techniques centered on total internal reflection fluorescence (TIRF), only the fluorophores (such as green fluorescent protein) in a thin subsurface layer of a target specimen are excited, resulting in more selective imaging. Instead of aiming a laser or other excitation light source at a specimen at the perpendicular, TIRF works by pointing a laser at a shallow angle along the surface of the target. If the light is at a certain threshold (the critical angle), the differences in refractive indices where the light meets the matter cause the light wave to bounce between the two surfaces without substantial loss (total internal reflection).
As a result of TIR, an evanescent wave forms and propagates just below the surface of the target, exciting any fluorophores present within the range of the evanescence field. Any possibly fluorescent particles outside of this range – only about 100 to 400 nm – remain unexcited.
Typically, the barrier between the coverslip and the imaging target – for example, a tissue sample with fluorescently tagged cell components – is where TIR happens and the evanescence field forms. The choice of area can be manipulated, but the surface of a sample is where most TIRF-based imaging takes place.

Epifluorescence microscopy and laser confocal scanning microscopy often are used in plant cell analysis but don’t offer the signal-to-background ratio of TIRF. Courtesy of the Journal of Experimental Botany.
In plant biology, the samples of most interest include various parts of roots, stems and leaves of larger vegetation. TIRF and the related technique, laser scanning confocal microscopy, frequently are used to study cellular dynamics, such as calcium-ion channel activity, in such samples. TIRF depends on the sample having the propensity to adhere to the underside of the coverslip, however, so that it remains within the evanescence field and, thus, the imaging area. Some potentially useful targets, such as the cells of various plants and fungi, either do not adhere well to coverslips or have thick walls that the evanescence field cannot penetrate well.
A few years ago, however, Sebastian Y. Bednarek and Catherine A. Konopka of the University of Wisconsin – Madison devised a method that they dubbed “variable-angle epifluorescence microscopy” (VAEM) to further reduce background noise and to address some of the disadvantages of TIRF.
Mechanically, the only difference between performing microscopy in TIRF or VAEM mode is the change in orientation of the mirrors in the TIRF instrument. Where TIRF uses a single, critical angle, VAEM calls for the use of a variety of subcritical angles. The major difference is that using subcritical angles in VAEM means that no evanescence field is generated, only a very thin band of illumination.
At the highest nonperpendicular, yet subcritical, angle, the field of illumination is very narrow, providing a high signal-to-noise ratio. The penetration depth they could achieve was as high as 700 nm.

Variable-angle epifluorescence microscopy (VAEM) uses illumination
beams directed at subcritical angles to light the target cell components
near the glass barrier. Total internal reflection fluorescence (TIRF)
microscopy, on the other hand, uses the critical angle to establish an
evanescence field that excites nearby fluorophores. Courtesy of The
Plant Journal.
Using an inverted microscope and TIRF attachment from Nikon, the researchers studied microtubule-binding domains in the root hairs of seedlings. They stepped the mirrors in the TIRF attachment through 10 decreasing angles of illumination and saw the illumination depth and average fluorescence intensity rise as the incidence angle decreased.
As with TIRF, the VAEM technique provided sharp images of fluorescently labeled structures, such as cytoskeletal and plasma membrane proteins. However, it also could peer more deeply and provide increased time resolution, thus improving the view of dynamic interactions.
Keeping an eye on the villins
Actin filament bundles are structures within the cytoskeletal array of plant cells. Bundling proteins such as villin collect individual actin filaments into these bundles, but the mechanism of action behind bundling remained elusive for a long time. Part of the problem was that there are several variants of villin, which may work together or independently of each other during the bundling process.
Christopher J. Staiger of Purdue University in West Lafayette, Ind., and his colleagues there and at the Commissariat á l’Energie Atomique in Grenoble, France, searched for a way to elucidate the role (or roles) of these villin variants in the commonly studied flowering plant Arabidopsis thaliana.
In one series of tests, Staiger’s group specifically tested the biochemical activities of two variants of villin – villin1 (VLN1) and villin3 (VLN3) – using TIRF microscopy and time-lapse imaging. To view the effect of VLN1 and VLN3 on bundling (including severing of existing bundles), the investigators used an Olympus inverted microscope outfitted with a Melles Griot 543-nm HeNe laser for excitation of the fluorophore on actin filaments and a Hamamatsu camera for detection.
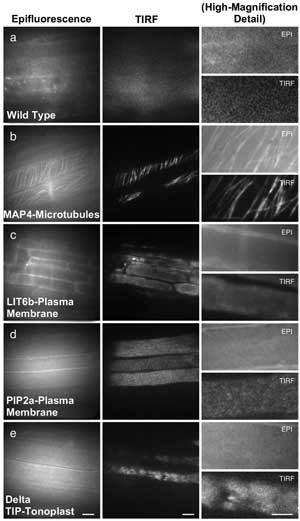
Epifluorescence microscopy and TIRF microscopy offer different levels of resolution in these examples from Arabidopsis thaliana. Scale = 10 µm in first two columns, 5 µm in the third column. Courtesy of the Journal of Experimental Botany.
Before either villin was introduced into the sample chamber, the rhodamine-actin filaments present barely moved, and few bundles were observed by the team. When either VLN1 or VLN3 was introduced, the investigators observed filaments finding each other and zipping together to form bundles. If calcium ions were present, however, VLN3 began to sever nearby bundles, decreasing filament length. The severing took place even when VLN1 was present as well.
To examine the formation of bundles in Arabidopsis epidermal cells, the group followed Bednarek and Konopka’s VAEM method, using an argon-ion laser from Melles Griot and the Hamamatsu camera with the same Olympus inverted microscope they used for TIRF. With this setup, they observed not only several filaments bundling into single structures, but also individual filaments participating in the creation of multiple bundles, all using the same zippering motion.
The group concluded that TIRF-driven research into the role of villins, especially VLN3, will help provide necessary information on the growth patterns and organization of cells in such plant organs and cells as roots and root hairs.
TIRF wars
VAEM was necessary because of TIRF microscopy’s perceived lack of ability to see past cell walls, which tend to be more rigid than mammalian cells, for example. Aiming a laser beam only at the critical angle generates an evanescence field too weak to pass through, but scanning through several subcritical angles overcomes this limitation, at least to an acceptable degree.
In the UK, Zoe A. Wilson and Gema Vizcay-Barrena of the University of Nottingham in Loughborough and their colleagues at Rutherford Appleton Laboratory in Didcot argue that TIRF microscopy is suitable not only for in vivo investigation of fluorescent proteins in plant cells, but also for single-molecule analysis.
Wilson’s group used a custom inverted TIRF microscope comprising a Zeiss objective lens, a laser made by Cobolt AB of Solna, Sweden, and a camera made by Andor Technology plc of Belfast, UK, to put TIRF and VAEM to the test.
Looking at intact parts of the roots of A. thaliana, the group found that fluorescently tagged particles could readily be imaged using TIRF microscopy in microtubules, nuclei and other organelles, as long as the steps taken to mount the samples were carefully performed. Ensuring, for example, that each root to be imaged touched the glass of the culture dish in which it rested was a strict requirement, to ascertain that it was within the evanescence field.
Comparing the two techniques, they found that the evanescence field generated during TIRF provides a better signal-to-background ratio (SBR) than does VAEM. They also observed that the higher SBR enables single-molecule imaging, in that deliberately photobleaching the majority of fluorophores present in a sample makes tracking the tagged protein of interest possible in high-SBR conditions.
Wilson and her colleagues ultimately argue that both TIRF and VAEM represent major advances for plant cell biology, but that TIRF’s higher SBR makes it more suitable for single-molecule analysis.