By incorporating techniques such as confocal, bright-field and wide-field imaging into high-content screening, researchers can now combine data previously locked away in separate assay and analysis methods.
KRIS VER DONCK, CONFLUENCE CONSULTING
Since the start of high-content screening (HCS) in the late 1990s, several imaging techniques have been applied for automated image capture, serving a variety of application goals and specifications. Two techniques known from related biological research fields were prominently present at the start: epi-fluorescence microscopy and laser-based cytometry. However, technological advancements are enabling HCS to cover new ground, particularly with confocal, bright-field and wide-field imaging.
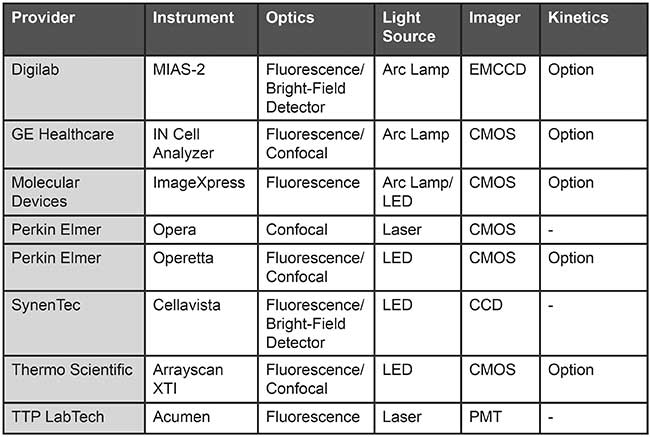
Table 1. An overview of automated high-content screening (HCS) plate readers. Based on information from provider websites.
Early HCS techniques
The combination of epi-fluorescence microscopy with targeted fluorescent probes in the 1950s provided an opportunity to spatially locate a molecular process in the cell. But laborious fixation and staining procedures only obtained static images frozen in time. The addition of cameras to manual microscope workstations provided the capability of storing images for later review, not for increasing sample capacity.
While a far cry from HCS as it is known today, this type of cellular imaging received a boost from the discovery of fluorescent proteins and the development of transfection probes to link reporter proteins to specific gene expression in living cells. These discoveries enabled researchers to study molecular processes in living cells and helped spur the development of microscope automation. In the mid- to late 1990s, PCs became fast enough to run such complex instruments, CCD cameras provided the speed to capture digital images and disk storage started to cope with the vast amounts of image storage from the first HCS screens.
In the 1960s flow cytometry was developed as a fast tool for diagnostic labs: Thousands of cells in suspension are moved one by one through a narrow flow cell. Response to the laser excitation of fluorescently tagged monoclonal antibodies are captured to fingerprint each cell. Adherent monolayer cells cannot be processed through a flow cell, but a 2D cytometry plot can be constructed on laser-scanning of a monolayer. Cells can be identified as responding cells versus nonresponding cells by the same cytometry classification techniques as used in flow. Although typically having lower spatial resolution than HCS microscopy, this technique can be extremely fast, enabling high-throughput screening assays.
Confocal imaging
Over time more techniques were added to improve HCS. For instance, laser scanning was combined with confocal optics to reduce background signals from outside the cell plane, while maintaining scan speed. Fluorescence microscopy was combined with confocal imaging to increase spatial resolution and allow for more detailed assays at the cost of speed and light yield. Several methods have been used to compensate for reduced light emission. For instance, on-chip image integration is a widely adapted method, with a prolonged image capture per frame that increases the sensitivity of the camera but slows down the imaging speed.
Increased laser light input can generate increased output and save reading time. But photobleaching of the fluorescent probe makes the sample unusable for repeated reading, as in kinetic live cell assays. Another approach is increased camera sensitivity. When the early noisy intensified photomultiplier cameras were replaced by electron multiplying CCD (EMCCD) cameras, speed and sensitivity became possible in combination with high image quality. These low light EMCCD techniques were added in 2007 to the MIAS-2 when Maia Scientific was acquired by Digilab Inc. Fluorescent light sources evolved from mercury or xenon lamps to longer-lasting laser sources and fast-switching LEDs. The shift from megapixel CCD cameras to larger CMOS cameras is gradually increasing the field of view (Table 1).
Bright-field applications
HCS is often used only in the context of fluorescent assays. But at the other end of the HCS spectrum, microscopy has returned to its roots with bright-field imaging for unstained cell cultures in applications where cell cultures are to be reused without external contamination from genetic transfection or chemical probes. In many HCS readers a form of bright-field imaging is implemented mostly for documentation purposes. It allows the visual review of the assay cultures similar to inspection of culture flasks in the cell culture lab. Only a few instruments other than the MIAS-2 have actually fully integrated the bright-field images into their screening analysis software applications.
Fluorescence image analysis is essentially as simple as identifying brighter objects in a dark background. But things become much more challenging on bright-field imaging. The first industrial scale application was found in the fully automated Cello robotic cell culture lab by The Automation Partnership in 2004. This robotic platform was designed to automate the clonal selection process in monoclonal antibody and protein production. The Cello incubators could house up to 900 plates in 384-well plates. A central robotic arm served two laminar flow cabinets for cell culture manipulation and a bright-field-only version of MIAS-2 served as a noninvasive cell culture plate monitoring reader. The entire culture monitoring process relied on fast bright-field imaging, down to five minutes per 96-well plate, combined with automated cell, colony and confluence quantification (Figure 1). In this way, multiple projects with well over 10,000 seedings per experiment could be completed in parallel in a time frame of about three weeks1.
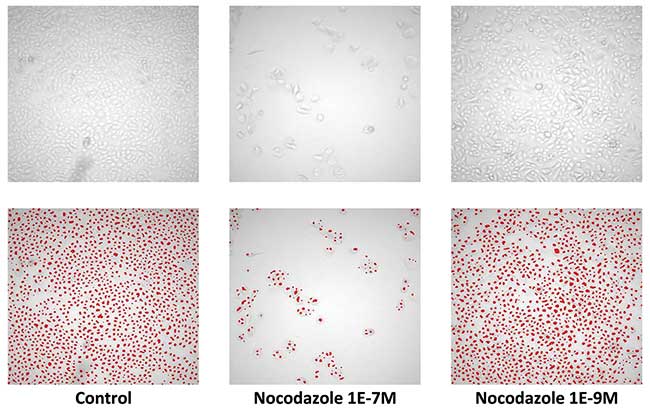
Figure 1. Unlabelled cell count on bright-field imaging. Image scanning with 5× objective on primary human keratinocytes. Cells seeded at low concentration and grown to confluence in control conditions (left) and grown following treatment with different concentrations of the microtubule inhibitor Nocodazole (middle and right). Top: brightfield images, bottom: cell detection analysis using eaZYX scale space detection algorithms for single cell identification. From Van Osta et al. (2002). The principles of scale space applied to structure and colour in light microscopy. Proceedings of the Royal Microscopical Society. Vol. 37, pp. 161-166.
Cell counting and label-free quantifications can also be combined as reference to other assays to improve the data quality (Figures 1, 2 and 3). Another bright-field example involved a visual stain that was used instead of fluorescence to report a BMP-2 induced muscle differentiation marker on a mesenchymal progenitor cell culture. Quantification of both muscle cells and differentiation marking relied entirely on bright-field imaging2.
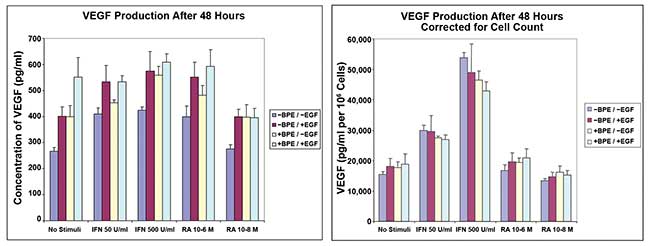
Figure 2. Vascular endothelial growth factor (VEGF) production in primary human keratinocytes, following IFNγ induction and retinoic acid (RA) modulation. VEGF endpoint ELISA (left) after 48 hours of incubation. VEGF endpoint ELISA (right) after 48 hours of incubation corrected with cell count on the same wells by bright-field imaging. Courtesy of Digilab Inc.
Field of view
Next to enabling applications and speed, another important consideration in HCS is capturing enough data to allow for statistical relevance of the screening output. In addition to providing data across thousands of sample wells in a screening, HCS can utilize the population statistics of the cell culture within each well. This requires the capturing of enough cells in each test condition. Several approaches are available to try and achieve this goal. The first approach is to have a large field of view.
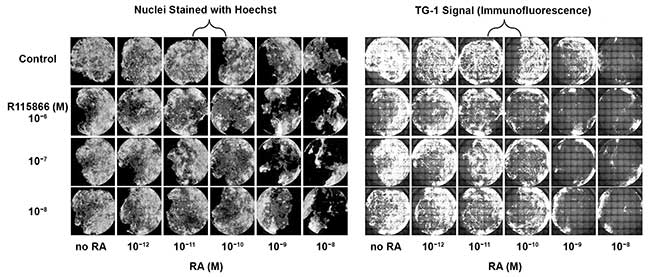
Figure 3. Differential response in human keratinocytes whole area of the wells scans of primary human keratinocytes in 96-well plates. Treatment with retinoic acid (RA) (left) results in a decrease in the number of nuclei (cells) present on the bottom of the wells, especially at higher concentrations. Upon treatment with RA (right), a dose dependent decrease in TG-1 signal (area and intensity) can be observed. The signal of cells treated with 10–10 M RA plus 10–6 or 10–7 M R115866 is lower as compared to cells treated with 10–10 M RA without compound. The signal with 10–9 M RA plus 10–8 M R115866 is lower than 10–9 RA without compound. The signal with 10–6 M R115866 without RA is lower as compared to nontreated cells. Courtesy of Digilab Inc.
With the right combination of a large imaging chip with a wide field-of-view objective, more cells can be captured in a single image. However, for certain applications, a minimum optical resolution is required to distinguish the features needed for the analysis. Higher resolution objectives, by definition, have a smaller field of view than low-resolution objectives. A 5× objective may be enough to visualize individual cells, but an objective of 20× or greater is often needed for many subcellular features. To compensate for the smaller field of view at higher magnification, multiple images can be sampled.
On some systems these images are independent subsampling in the well. However, many systems have the option to combine multiple images into a super-image. Often the technique of image joining by overlap is used to create a super-image, which involves a feature-mapping algorithm that looks at two overlapping adjacent images and joins them on the overlapping positions of objects in both images. Although these matching algorithms work fine, they come at a cost of speed because each image pair needs to overlap by about 20 to 30 percent. Some higher resolution systems are equipped with high-precision XY motor assemblies to join images into a super-image by mechanical positioning of the subsampling images. They scan the defined area tile by tile and join the images without requiring image overlap. This technique was applied in the MIAS-2 for the Cello robot, scanning entire wells in 6-well plates to monitor the total cell count in a clonal culture expansion.
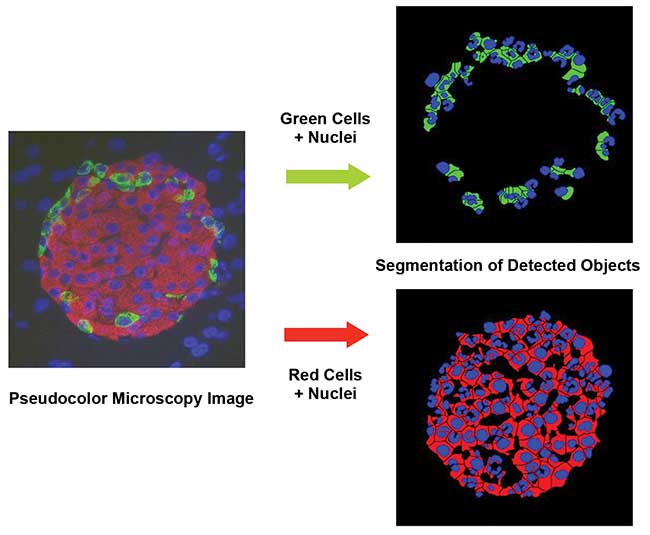
Figure 4. A three-color stained image with differential immunomarker labeling. Analysis by the eaZYX Color Scale-Space software. Courtesy of Digilab Inc.
3D
A large field of view does not have to stop at 2D images and can be expanded into the third dimension. In confocal, the Z-stack is used to build a 3D construction image of subcellular structures. The confocal HCS devices all feature a form of Z-stacking. But researchers can also apply Z-stacks for screening 3D culture systems or cell clusters like (Figure 1). Screens on cell clusters in a gel culture have been used in the search for potential tumor treatments.
In the ideal case, extended scans in the three spatial dimensions can be combined with time series on systems with live cell or incubator options. There are also advantages to adding multiple imaging modes, such as fluorescence and bright-field. Most HCS instruments offer subsets of these features due to their taxing effect on speed. Only a few, such as the MIAS-2, are capable of combining all, leaving the users the choice of what scanning combination they need for their screening project.
Growth potential
The HCS field will grow beyond its current boundaries by evolutions that have already started. New types of cameras can be expected to enter into HCS. One particularly promising evolution is the development of hyperspectral cameras. Rather than taking a three-color, red-green-blue image, the hyperspectral camera can take complete wavelength spectra for each spatial pixel. The result is not a 2D color image but a dense 3D data structure. This hyperspectral dataset may support improved analysis of color-stained tissues or of multilabel fluorescent assays in the future.
The data storage capacity on file servers has increased well into the terabyte ranges and continues to grow. With recent evolutions of accessing vast datasets in the cloud, researchers are now building access to “Big Data,” not just on the image storage but also on image feature extraction data. The sheer volume of images and image-derived data can be expected to feed large-scale statics analysis beyond what has been feasible.
Another effect of this evolution already noticeable on smartphones is the intelligent photo app, which recognizes individual people from images with increasing fidelity. The analysis paradigms behind this image recognition are being created by using the big data for a deep learning approach on image feature extractions to identify which combinations of feature extractions lead to trained analysis paradigms.
Once such paradigms are defined, typically any computer will be powerful enough to quickly run this image recognition. This image analysis revolution is driven by a continuous stream of pictures available in the cloud and will result in new image analysis paradigms that can benefit HCS. In the end, it will be the creativity of the screening-application developer to decide how to apply these new enabling techniques to the benefit of new disease insights and the development of treatments.
Meet the author
Kris Ver Donck is an automation technology consultant at Confluence Consulting BVBA in Herent, Belgium; email [email protected].
References
1. K. Lindren et al. (January 2009). Automation of cell line development. Cytotechnology, Vol. 59, pp. 1-10.
2. Darcy et al. (June 2012). Bone, Vol. 50, pp. 1294-1303.