Nature’s precise structural organization inspires the development of advanced probes that amplify weak Raman signals, strengthening cancer detection.
SWATI TANWAR AND ISHAN BARMAN, JOHNS HOPKINS UNIVERSITY
In biomedical applications, in which the target analyte is often at nanomolar or picomolar concentrations, the effectiveness of Raman spectroscopy is limited. This is due to the inherently weak Raman effect, where typically only one in 10 million photons is inelastically scattered. Furthermore, in biological milieus, distinguishing the vibrational signatures of target molecules from other Raman-active molecules can be challenging. Beyond the conventional label-free modality, several strategies have been adopted to overcome the limitations of the inherently weak Raman effect, of which surface-enhanced Raman spectroscopy (SERS) remains the most widely used. By leveraging the electromagnetic field enhancement provided by plasmonic nanoparticles, SERS can increase Raman scattering intensity by many times. However, persisting challenges, such as the nonuniform generation of hotspots (localized regions of intense enhancement) leading to non-reproducible amplification of Raman signals, make quantitative analysis challenging.
Nanoprobes equipped with Raman
reporters illuminate intricate biological
structures, pushing the boundaries of
Raman spectroscopy. Courtesy of Arnab
Chatterjee/Barman Laboratory at Johns
Hopkins University.
Raman spectroscopy, renowned for its exceptional molecular specificity and noninvasive analytical prowess, has long been valuable in biology, physics, and industry. While the Raman effect was discovered by C.V. Raman in 1928, its applicability for practical applications was realized only after the advent of lasers in 1960. This advancement, complemented by subsequent developments, such as charge-coupled devices (CCDs) and enhanced optical filters, enabled the design of Raman-specific systems. It was soon appreciated that Raman spectroscopy, with its unique ability to elucidate molecular compositions, would emerge as an indispensable analytical tool with broad applicability, revolutionizing diverse applications ranging from industrial processes to clinical diagnostics. However, the technique’s efficacy is predominantly evident in scenarios in which the material being analyzed is present in bulk. Trace samples are problematic for this modality, and it has taken innovative engineering approaches to enhance its effectiveness in this situation in a clinical context.
By leveraging the electromagnetic field enhancement provided by plasmonic nanoparticles, SERS can increase Raman scattering intensity by many times.
The frontier of Raman spectroscopy applications has expanded with the development of intelligent optical nanoprobes, engineered by leveraging the principles of self-assembly. Self-assembly, a strategy observed in nature for generating functional structures, involves assembling smaller units into structural hierarchies at various scales. These structures exhibit precise organization, shape, and functionalities. Drawing inspiration from nature’s design and chemistry, engineers of SERS technology combine principles from biology, chemistry, and engineering to create Raman-active optical probes (opening image). The goal of such work is to amplify weak signals, enhance the detection of low-concentration analytes, and address the challenge of signal reproducibility in complex biological environments. DNA has emerged as a preferable choice as the basis of such probes, because it offers high programmability and efficiency in assembly into biocompatible optical materials.
DNA-assembled nanoprobes
DNA, traditionally recognized for carrying genetic information across generations, has emerged as a superior building block for advanced structured materials through the process of self-assembly. The technique relies on the hybridization of complementary single-stranded DNA sequences into double-helical structures. Due to its sequence specificity and the vast potential for designing bespoke sequences, DNA offers a richly versatile tool kit for creating complex optical structures.
Recently, attention has been diverted to merging DNA nanotechnology with plasmonics1. This innovative combination is crucial for advancing SERS-based optical nanoprobes by using DNA technology to integrate spatial positioning into configurable single-stranded DNA elements. This capability is essential for precisely arranging plasmonic particles into specific, gap-controlled configurations, significantly enhancing the reproducibility of SERS signals and broadening their potential applications in various fields.
Among various SERS-active optical structures, optical cavities are potentially very useful due to their ability to confine light within deep-subwavelength regions (a few to tens of nanometers). This effect creates intense local electromagnetic fields, which are highly desirable for single-molecule SERS studies. However, fabricating structurally resilient hierarchical plasmonic nanocavities with high fidelity using traditional lithography methods remains a profound challenge. These nanocavities are difficult to produce even using DNA-based self-assembly, given that optical cavity structures typically require the precise co-assembly of multiple optical components into a large, rigid structure for maximum enhancement. And DNA is inherently flexible, which can compromise the structural resiliency and precise alignment needed in the cavity regions for optimal functionality.
DNA-silicified template
Recently, a novel method has been developed to construct structurally resilient plasmonic nanocavities, termed DNA-silicified template for Raman optical beacon (DNA-STROBE) (Figure 1)2. This technique introduces an original approach versus traditional methods, by merging the precision, programmability, and nanogap-control capabilities of DNA with the chemical inertness and structural stability of silica. Silica, an inert inorganic material, not only exhibits excellent biocompatibility and nontoxicity but also provides thermal and chemical stability. These properties make it an ideal candidate for biomaterial encapsulation.
In the DNA-STROBE method, hierarchical DNA nanoassemblies are initially formed using unique nitrogen base-pairing interactions, and subsequently coated with a thin layer of amorphous silica via DNA-templated sol-gel chemistry (Figure 1a). This encapsulation process effectively stabilizes the DNA within an ultrathin silica shell, preserving its structural integrity. This method addresses the practical difficulties of electrostatic repulsion between the anionic DNA phosphate and silanol precursors in the silica coating solution. To overcome this, positively charged N-trimethoxysilylpropyl-N,N,N-trimethylammonium chloride (TMAPS) is employed as a co-structure directing agent. TMAPS facilitates direct co-condensation at the sites required for silica shell growth on the DNA constructs. The resulting DNA-STROBE constructs demonstrate significant chemical inertness and structural stability over a broad range of pH values, effectively condensing the nanogaps to produce an array of sub-nanometer plasmonic cavities (Figure 1b).
Leveraging the ultrasmall mode volume of the DNA-STROBE constructs, the method facilitated single-molecule SERS-based plasmonic sensing. Employing principles of molecular diffusion into and out of these mode volumes, along with superresolution imaging techniques, precise localization of distributed SERS hotspots beyond the diffraction limit was achieved (Figure 1c). Unlike techniques that manipulate individual molecules directly, this approach allowed probing single-molecule activities within higher solution concentrations, up to the micromolar range.
This capability offers a unique opportunity to noninvasively delve into the mechanics of biological processes that were previously considered out of reach. The adaptability of the DNA-STROBE method supports the fabrication of a diverse array of biomimetic metal-silica hybrid materials, extending the foundational configuration to a variety of applications, such as biosensing, monitoring physiological metabolism, and evaluating the effectiveness of drug candidates. The highly programmable and reproducible nature of DNA-STROBE, combined with its quantitative, label-free molecular analysis capabilities, provides Raman spectroscopy with a powerful and generalizable tool for exploring both geometric and molecular properties.
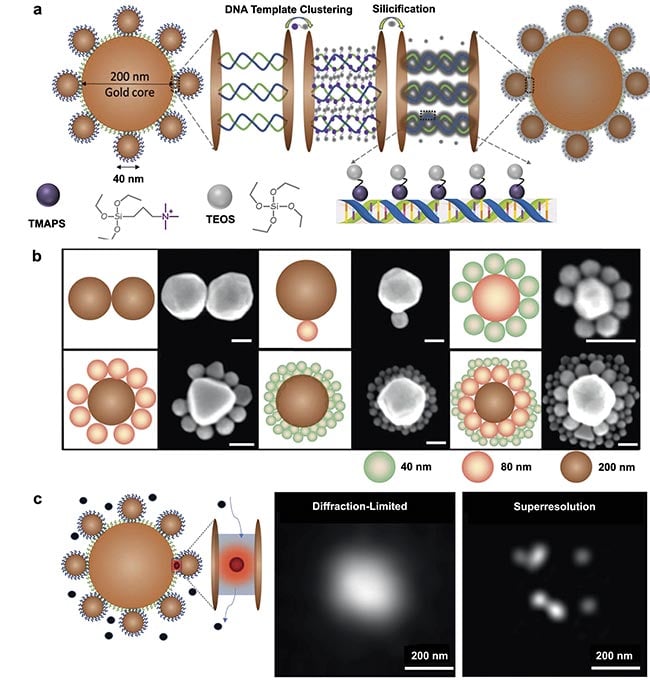
Figure 1. A schematic illustration of a DNA-silicified template for Raman optical beacon (DNA-STROBE) design and fabrication (a). Schematics
and scanning electron microscopy images of gold nanodimers, core-satellite and core-satellite-satellite nanoassemblies (scale bar =
100 nm) (b). Standard diffraction-limited imaging contrasted with single-molecule, superresolution surface-enhanced Raman spectroscopy
(SERS) measurements using DNA-STROBE (c). Adapted with permission from Reference 2.
Plasmonic nanoprobe targets cancer cells
Building on the sophisticated capabilities inherent in the DNA origami technique — an advanced evolution of standard DNA-based assembly methods that is sequence programmable — the SERS-enabled plasmonically coupled nanoprobe (SPECTRA) represents a significant advancement in targeted cancer imaging (Figure 2)3. Using the precision folding of single-stranded DNA into complex nanostructures, this technique facilitates the controlled assembly of metallic nanoparticles with precise nanoscale accuracy. Such architecture allows for optimal positioning of nanoparticles to enhance plasmonic coupling effects, significantly amplifying Raman signals for heightened sensitivity.
Figure 2. This schematic illustrates the use of surface-enhanced Raman spectroscopy (SERS)-enabled plasmonically coupled nanoprobe
(SPECTRA) for targeted Raman imaging in detecting metastatic DU145 prostate cancer cells. SPECTRA’s effectiveness is underscored by the
prominent nitrile peak at 2225 cm−1, primarily detected in DU145 cells due to SPECTRA’s higher affinity for these specific cells. Adapted with
permission from Reference 3.
At the core of SPECTRA’s design lies a dimer configuration of identical gold nanorods, meticulously aligned end-to-end on a rectangular DNA origami template. This strategic arrangement minimizes interparticle gaps to create a highly sensitive plasmonic nanoantenna. The template is further functionalized with a specific DNA aptamer sequence targeting metastatic prostate cancer DU145 cells, offering remarkable specificity. Additionally, the integration of a Raman reporter molecule, 4-mercaptobenzonitrile (4-MBN), chosen for its vibrational frequency within the cell-silent Raman window, enables SPECTRA to produce a distinct and strong nitrile signal at 2225 cm−1, facilitating clear differentiation between cancerous and noncancerous cells.
The multifunctional nature of the DNA origami template enables the assembly of multiple components, including nanorods in an end-to-end configuration, a cancer-targeting DNA aptamer, and 4-MBN on one optical beacon to perform multiple functions, illustrating the versatility and potential of this technique for creating complex, versatile nanodevices in a highly controlled manner (Figure 3a).
The effectiveness of SPECTRA in distinguishing cancer cells was validated through rigorous in vitro Raman imaging studies with DU145 and LNCaP prostate cancer cells. Incubation with SPECTRA resulted in DU145 cells exhibiting a significantly higher intensity of the nitrile signal compared with LNCaP cells, underscoring the probe’s remarkable targeting capabilities (Figure 3b). Notably, the occupancy ratio of SPECTRA within the total cell area was fivefold higher for DU145 than for LNCaP, demonstrating the probe’s superior ability to differentiate between metastatic cancer cells and those with lower metastatic potential. This could aid in tumor margin assessment and guided biopsies.
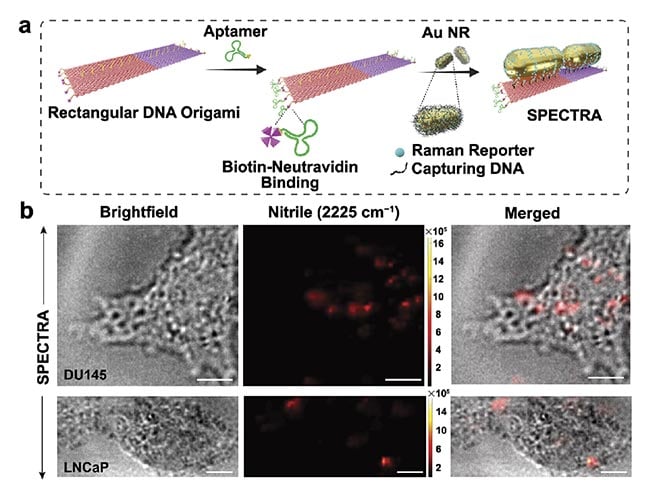
Figure 3. The step-by-step fabrication of the surface-enhanced
Raman spectroscopy (SERS)-enabled plasmonically coupled
nanoprobe (SPECTRA). An aptamer sequence specific to DU145
cells was selectively attached to the edges of a rectangular
DNA origami template via biotin-neutravidin conjugation. Gold
nanorods labeled with 4-mercaptobenzonitrile (4-MBN) were then
arranged on the template in a nanoantenna configuration (a). In
vitro cellular Raman imaging of DU145 and LNCaP cells incubated
with SPECTRA (scale bar = 10 μm) (b). Adapted with permission
from Reference 3.
The enhanced detection sensitivity of SPECTRA, highlighted by an enhancement factor calculation, revealed a three-order magnitude increase in sensitivity compared with conventional gold nanorod monomers. This translates into the capability of detecting lower concentrations of target cells. As a pioneering innovation in biophotonics and molecular imaging, SPECTRA exemplifies the integration of advanced DNA origami with plasmonic nanotechnology, providing a highly specific and sensitive tool for the early detection of metastatic cancer cells. The scalability and reproducibility of the DNA origami technique further enhances SPECTRA’s clinical use, marking a new era in precision diagnostics. These cellular models will need to be tested on animal specimens before advancing to human trials.
Future promise in medicine
During the past few decades, Raman spectroscopy has seen immense advancements, driven by groundbreaking developments in detection technologies and probe design. These innovations have catalyzed a surge in research, aiming to harness the technique for a spectrum of clinical applications, from diagnostics to surgical interventions. Yet, the full clinical potential of Raman spectroscopy hinges on overcoming significant challenges through the development of advanced optical nanoprobes.
At the forefront of this scientific evolution is the integration of DNA nanotechnology. This approach has revolutionized the creation of multifunctional SERS-based nanoprobes. Using DNA-based self-assembly methods, optical devices capable of precise and targeted functionalities have been crafted, which are crucial for addressing complex medical diagnostics. The path forward will require an amalgamation of innovative strategies in chemistry and materials science to enhance the functionality, stability, and application of these nanoprobes. The continuous refinement of DNA nanotechnology promises to push the boundaries of what is possible with Raman spectroscopy in medicine, potentially transforming how clinicians can detect and manage disease.
Meet the authors
Swati Tanwar is a postdoctoral fellow in the Department of Mechanical Engineering at Johns Hopkins University. Specializing in nanotechnology and synthesis chemistry, her research focuses on developing innovative nanodevices using nature’s tool kit — DNA and peptides — for monitoring biological processes and diagnostic applications. She has developed Raman-active, stimuli-responsive nanoprobes to improve targeted cancer imaging, detection, and drug delivery; email: [email protected].
Ishan Barman, a professor of mechanical engineering with joint appointments in the Johns Hopkins Sidney Kimmel Cancer Center and the Russell H. Morgan Department of Radiology and Radiological Science, is an expert in biomedical imaging and bioengineering. His research program is dedicated to engineering innovation for the analysis of complex biological systems. His work involves engineering nanostructured probes for the ultrasensitive detection of specific molecular species through SERS, capturing intricate tissue changes predicting disease progression. Another focus is quantitative tissue microspectroscopy, aiming to identify differential spectral markers for cell types, early cancer detection, and objective cancer grading; email: [email protected].
References
1. N. Liu and T. Liedl. (2018). DNA-assembled advanced plasmonic architectures. Chem Rev, Vol. 118, No. 6, pp. 3032-3053.
2. L. Liang et al. (2021). A programmable DNA-silicification-based nanocavity for single-molecule plasmonic sensing. Adv Mater, Vol. 33, No. 7, p. 2005133.
3. L. Wu et al. (2024). DNA origami-engineered plasmonic nanoprobes for targeted cancer imaging. Adv Funct Mater, Vol. 34, No. 30, p. 2309929.